Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.


ments
can act
only
on transcription
units
within the
same domain.
A
domain
might
con-
tain
more than
one transcription
unit and/or
enhancer.
FiGUR[
8S"55
summarizes
the
structures
that
might be involved
in
defining
a
domain.
An insulator
stops
activating
or repress-
ing
effects from passing.
In
its
simplest form,
an insulator
blocks
either
type
of effect
from
passing
across
it, but
there
can be more
com-
plex
relationships
in which
the
insulator
blocks
only one type
of effect
and/or
acts
direction-
ally.
We assume
that insulators
act by affect-
ing higher-order
chromatin
structure,
but we
do not know
the details
and
varieties
of such
effects.
A matrix
attachment
site
(MAR)
may
[re
responsible
for
attaching
chromatin
to a
site on
the nuclear
periphery
(see
Section 28.6,
Spe-
cific
Sequences Attach
DNA
to an
Interphase
Matrix).
These
are likely
to be responsible
for
creating
physical
domains
of DNA
that take the
form
of loops
extending
out from
the attach-
ment
sites. This looks
very much
like
one model
for insulator
action. In
fact,
some MAR
elements
behave as insulators
in
assays invitro,butil
seems that their
ability
to atrach
DNA to
the
matrix
can be separated
from
the insulator
func-
tion,
so there is not
a simple
cause and
effect.
It would not
be surprising
if insulator
and MAR
elements
were associated
to maintain
a rela-
tionship between
regulatory
effects
and
phys-
ical
structure.
An LCR
functions
at a distance
and may
be
required
for any
and all
genes
in
a domain to
be expressed
(see
Section
29.20,
An LCR May
Control
a Domain).
When a
domain has an LCR,
its
function is
essential for
all
genes
in the
domain,
but LCRs do not
seem
to be common.
Several types
of cis-acting
structures
could be
required
for function.
As defined
originally,
the
property
of
the LCR
rests
with an
enhancer-like
hypersensitive
site that
is needed
for the full
activity
of
promoter(s)
within
the domain.
The organization
of domains
may help
to
explain the large
size
of the
genome.
A certain
amount of space
could be required
for
such a
structure to
operate, for
example, to allow
chro-
matin
to become decondensed
and
to beconte
accessible.
Although
the exact
sequences of
much
of the unit
might be irrelevant,
there
might
be selection for the
overall
amount of
DNA
within it, or at least
selection
might
pre-
vent
the various transcription
units from
becom-
ing too
closely spaced.
nsulator MAR
LCR Enhancer
t-r{;7
Transcription units
flGllR*
?*"55 Domains
may
possess
three types of sites:
insulators
to
prevent
effects
from
spreading between
domains, MARs
to attach
the domain
to the nuclear
matrix.
and
LCRs
that are
required for ini-
tiation oftranscription. An
enhancer
may act on more than one
pro-
moter within
the domain.
Summary
All eukaryotic
chromatin
consists of nucleo-
somes.
A nucleosome contains a characteristic
length
of DNA, usually
-200
bp, which is
wrapped around an octamer containing two
copies each
of
histones H2A,H2B, H3, and H4.
A single Hl
protein
is associated with each
nucleosome.
Virtually all
genomic
DNA is
orga-
nized into nucleosomes. Tfeatment with micro-
coccal
nuclease shows that
the DNA
packaged
into
each nucleosome can be divided opera-
tionally into two regions. The
linker region is
digested rapidly
by the
nuclease; the core region
of 146
bp
is
resistant
to
digestion.
Histones H3
and H4 are the most highly conserved,
and an
H32-H42tetramer
accounts
for the diameter of
the
particle.
The H2A and H2B histones are
organized as two H2A-H2B dimers.
Octamers
are assembled by the successive
addition of two
H2A-H2B dimers to the H3z-H42
kernel.
The
path
of
DNA around the histone
octamer creates
-1.65
supercoils.
The DNA
"enters"
and
"leaves"
the nucleosome
in tne
same
vicinity, and could
be
"sealed"
by
histone
Hl.
Removal of the core
histones releases
-L0
supercoils. The difference can
be largely
explained by a change
in the helical
pitch
of
DNA, from an average of 10.2 bp/turn
in nucle-
osomal form to 10.5 bp/turn
when free
in
solu-
tion. There is variation
in the structure of DNA
from a
periodicity
of 10.0 bp/turn
at the nucle-
osome
ends to
10.7 bp/turn in the center.
There
are kinks
in the
path
of
DNA on the
nucleosome.
Nucleosomes
are organized
into a fiber of
30 nm
diameter
that has six
nucleosomes
per
turn and
a
packing
ratio of
40. Removal of Hl
allows this fiber to unfold
into a
l0 nm fiber
that consists of a linear string
of nucleosomes.
The l0 nm fiber
probably
consists
of the l0 nm
fiber
wound into a
2-start solenoid.
The l0 nm
fiber is the
basic
constituent
of both euchro-
matin and heterochromatin;
nonhistone
29.22 Summary
791

proteins
are responsible
for further organiza-
tion
of the
fiber into chromatin or chromosome
ultrastructure.
There are two
pathways
for nucleosome
assembly. In the replication-coupled
pathway,
the
PCNA
processivity
subunit of the
replisome
recruits CAF-1, which
is
a
nucleosome as-
sembly factor. CAF- I assists the deposition
of
H32-H42
tetramers
onto the daughter duplexes
resulting from replication. The tetramers
may
be
produced
either by disruption
of existing
nu-
cleosomes by the replication fork or as the
result
of assembly from newly synthesized
histones.
Similar sources
provide
the H2A-H2B dimers
that then assemble with the
H32-H42tetramer to
complete the nucleosome. The H3z-H4ztetramer
and the H2A-H2B dimers assemble at random,
so the
new nucleosomes may include both
pre-
existing and newly synthesized
histones.
RNA
polymerase
displaces histone octamers
during transcription.
Nucleosomes
reform on
DNA
after the
polymerase
has
passed,
unless
transcription is very intensive
(such
as
in rDNA)
when
they
may
be displaced completely.
The
replication-independent
pathway
for nucleo-
some assembly
is responsible for replacing his-
tone octamers that
have
been displaced
by
transcription. It uses the histone
variant H3.l
instead of Hl. A similar
pathway,
with another
alternative
to
H3, is
used
for
assembling
nu-
cleosomes at centromeric DNA sequences
fol-
lowing replication.
An insulator blocks the transmission of acti-
vating
or
inactivating
effects
in
chromatin.
An
insulator
that
is located
between an enhancer
and a
promoter
prevents
the enhancer from
activating the
promoter.
T\ryo insulators define
the region between them as a regulatory
domain; regulatory interactions within the
domain are limited to it, and the domain is insu-
lated
from outside effects. Most insulators block
regulatory
effects
from
passing
in either direc-
tion, but some are directional. Insulators usu-
ally can block
both
activating
effects
(enhancer-
promoter
interactions) and inactivating effects
(mediated
by spread
of
heterochromatin),
but
some are limited
to one or the other. Insulators
are thought to act via
changing
higher-order
chromatin
structure, but the details are not
certaln.
TWo
tlpes of changes in sensitivity to nucle-
ases
are associated with
gene
activity. Chro-
matin
capable of being transcribed has a
generally
increased sensitivity
to
DNAase I,
reflecting
a change in structure
over
an exten-
sive region that
can be defined as a domain
con-
taining
active or
potentially
active
genes.
Hlper-
sensitive sites
in DNA occur at discrete
locations,
and are
identified by
greatly
increased
sensitiv-
ity to DNAase L
A hypersensitive site consists
of a sequence
of
-200
bp
from
which
nucleo-
somes
are excluded by
the
presence
of other
proteins.
A hypersensitive site
forms
a bound-
ary that
may cause adjacent
nucleosomes
to be
restricted
in
position.
Nucleosome
positioning
may be important
in controlling access of reg-
ulatory
proteins
to DNA.
Hypersensitive sites occur at several types
of regulators.
Those that regulate transcription
include
promoters,
enhancers, and
LCRs.
Other
sites include origins
for replication and cen-
tromeres. A
promoter
or enhancer acts on a sin-
gle gene,
but an
LCR
contains a
group
of
hypersensitive sites
and may regulate a domain
containing several
genes.
References
The Nucleosome Is
the Subunit
of A[[ Chromatin
Reviews
I(ornberg, R.
D.
(1977).
Structure of chromatin.
Annu. Rev. Biochem.
46,9)l-954.
McGhee, J. D., and Felsenfeld, G
(1980).
Nucleo-
some structure.
Annu. Rev. Biochem 49,
ltlS-t156.
Resea rc h
I(ornberg,
R. D.
(1974).
Chromatin structure: a
repeating unit of
histones
and
DNA.
Science
184, 868-87 r.
Richmond, T.
J.,
Finch, J. T., Rushton, B.,
Rhodes, D., and
Klug, A.
(1984).
Structure ot
the
nucleosome
core
particle
at
7 A resolu-
tion. Nature
Jll, 5j2-5)7 .
DNA Is
Coited
in Arravs
of
Nucteosomes
Rese a
rc h
Finch,
J.
T. et al.
.1977).
Structure
of
nucleosome
core
particles
of chromatin . Nature 269,
29-36.
Nucteosomes Have a Common
Structure
R esea
rc h
Shen, X. et al.
(I995).
Linker histones
are not
essential and affect chromatin
condensation
in vitro. Cell 82, 47-56.
792 CHAPTER 29
Nucleosomes

DNA
Structure
Varies
on
the NucteosomaI
Surface
Review
Wang, J.
(1982\.
The
path
of DNA
in
rhe nucleo-
some.
Cell 29, 724-726.
Res ea rc h
Richmond, T.
J. and Davey,
C. A.
(2003).
The
structure
of
DNA
in
the nucleosome
core.
Nature 42j,
145-150.
The Periodicity
of DNA
Changes
on the Nucteosome
Review
Travers,
A. A. and Klug,
A.
(19871.
The
bending
of DNA in
nucleosomes
and
its wider implica-
tions. Philos Trans.
R.
Soc
Lond
B Biol.
Sci.317.
5)7-561.
0rganization
of the Histone
Octamer
Resea rc h
Angelov,
D., Vitolo,
J. M., Mutskov
V., Dimitrov
S.,
and Hayes,
J. J.
(2001
).
Preferential
interac-
tion of the core histone
tail domains
with
Iinker
DNA. Proc
Natl Acad.
Sci.
USA98,
6599-6604.
Arents,
G.,
Burlingame,
R. W.,
Wang, B.-C., Love,
W. E., and Moudrianakis,
E.
N.
(1991).
The
nucleosomal
core histone
octamer
at 3I A res-
olution: a
tripartite
protein
assembly
and a
left-handed
superhelix. Proc
Natl. Acad. Sci
us.4 88,
r0148-10r52.
Luger, I(.
et al.
(1997).
Crystal
structure of
the
nucleosome
core
parricle
at 28 A resolution.
Nature 389,251-260.
The Path
of Nucteosomes
in the
Chromatin Fiber
Review
Felsenfeld,
G.
and
McGhee,
J. D.
(1986).
Srrucrure
of the 30 nm
chromatin
fiber.
Cell
44,
375-]77.
Resea rch
Dorigo, B.,
Schalch, T., I(ulangara,
A.,
Duda, S.,
Schroeder,
R. R., and Richmond,
T.
J.
(2004).
Nucleosome arrays
reveal the
two-start orga-
nization
of the chromatin
fiber.
Science
jO6.
t57 t-l573.
Schalch, T.,
Duda, S.,
Sargent, D. F.,
and Rich-
mond, T. J.
(2005).
X-ray structure
of a
tetranucleosome
and its implications
for
the
chromatin
Libre. Nature 4)6,
lJ8-141.
Reproduction
of Chromatin
Requires
Assembty
of
Nucteosomes
Osley, M. A.
(1991).
The
regulation
of histone
syn-
thesis in
the cell cycle. Annu.
Rev. Biochem
60,
827-86t.
Reviews
Verreault,
A.
(2000).
De novo nucleosome assem-
bly: new
pieces
in an old
puzzle.
Genes Dev. 14,
t4)o-14]8.
Resea rc h
Ahmad,
I(. and Henikoff, S.
(2001).
Centromeres
are specialized replication domains in hete-
rochromatin.
J
Cell
Biol l5 3. l0l-l 10.
Ahmad,
I(.
and
Henikoff,
S.
(2002).
The histone
variant H3.3 marks active chromatin by
replication-independent
nucleosome
assembly. Mol.
Cell
9, ll9l-I200.
Gruss,
C., Wu, J.,
I(oller, T.,
and Sogo, J.
M.
(19931
. Disruption of the
nucleosomes at
the
replication
fork. EMBO J.
12, 453)-4545.
Loppin,
B., Bonnefoy, E., Anselme, C., Laurencon, A.,
I(arr, T. L.,
and Couble,
P.
(2005).
The histone
H3.3
chaperone HIRA
is
essential
for chro-
matin assembly in the male
pronucleus.
Nature 437, lj86-l)90.
Ray-Gallet,
D.,
Quivy,
J. P., Scamps, C.,
Martini,
E. M., Lipinski, M., and Almouzni, G.
(2002).
HIRA is
critical
for a nucleosome assembly
pathway
independent of DNA synthesis. Mol.
Cell9, 109l-1100.
Shibahara,
I(., and Stillman,
B.
(19991.
Replication-dependent marking of DNA by
PCNA facilitates CAF- I
-coupled
inheritance
of
chromatin.
Cell
96, 57 5-585.
Smith, S. and Stillman, B.
(1989).
Purification and
characterization of CAF-I,
a human cell factor
required
lor chromatin assembly
during DNA
replication in vitro. Cell 58,
15-25.
Smith, S. and
Stillman,
B.
(1991).
Stepwise
assem-
bly
of chromatin
during DNA
replication
in
vitro.
EMBO J. lO,
97 1-980.
Tagami, H.,
Ray-Gallet,
D., Almouzni, G., and
Nakatani, Y.
(2004).
Histone Hl.I and Hl.l
complexes mediate nucleosome
assembly
pathways
dependent or
independent of DNA
synthesis.
Cell
ll6, 5l-61.
Yu, L.
and Gorovsky,
M. A.
(1997).
Constitutive
expression, not a
particular primary
sequence,
is
the important feature
of the H3 replacement
variant
Llv2 in Tetrahymena
thermophila. Mol.
Cell. Biol 17, 6]03-6310.
Are Transcribed
Genes
0rganized
in Nucteosomes?
Review
I(ornberg,
R. D. and Lorch,
Y.
(1992).
Chromatin
structure and transcriprion.
Annu. Rev. Cell
Biol 8, 56)-587.
Histone
Octamers
Are
Disptaced
by Transcription
Researc h
Cavalli,
G. and
Thoma, F.
(
1993
)
.
Chromatin
tran-
sitions during activation
and repression of
galactose-regulated genes in
yeast.
EMBO J.
12,460)46r).
References 793

@
Reviews
Studitsky,
V.
M., Clark, D. J., and
Felsenfeld, G.
(1994).
A histone octamer can step around
a
transcribing
polymerase
without
leaving the
remolare. cell
7 6. 37 l-382.
@
Nucleosome Disp[acement and
Reassembty Require SpeciaI
Factors
Resea rc h
Belotserkovskaya, R., Oh, S., Bondarenko, V.
A.,
Orphanides, G., Studitsky,
V. M., and Rein-
berg, D.
(2001).
FACT
facilitates
transcription-
dependent nucleosome alteration. Science 3Ol,
1090-1093.
Saunders, A., Werner, J., Andrulis, E. D.,
Nakayama,
T., Hirose, S., Reinberg, D., and
Lis,
J.
T
(2003).
Tracking FACT and the RNA
polymerase
II
elongation complex through
chromatin in vivo. Science )Ol,
1094-1096.
Insulators Btock the
Actions
of
Enhancers
and
Heterochromatin
Gerasimova, T. I. and Corces, V. G.
(2001).
Chro-
matin insulators and boundaries: effects on
tra
nscription
and
nu
clear or
ganizalion.
An n u.
Rev.
Genet. 35,
19)-208.
West, A.
G., Gaszner,
M.,
and
Felsenfeld,
G.
(20021.
Insulators:
many functions, many
mechanisms. Genes Dev. 16. 27 l-288.
?W
Insulators
Can
Define
a
Domain
Resea
rc
h
Chung, J. H., Whiteley, M., and Felsenfeld, G.
(19%l
. A 5' element of the chicken
p-globin
domain serves as an insulator in human ery-
throid
cells and
protects
against
position
effect
rn Drosophila
Cell
7 4,
505-514.
Cuvier,
O.,
Hart,
C.
M.,
and
Laemmli,
U.
K.
(1998).
Identification of a class of chromatin bound-
ary elements. Mol.
Cell
Biol 18,7478-7486.
Gaszner, M.,Yazquez,
J.,
and
Schedl,
P.
(1999).
The Zw5
protein,
a component of the scs
chromatin domain boundary, is able to block
enhancer-promoter interaction.
Genes
Dev. l),
2098-2107.
I(ellum,
R. and Schedl, P.
(
I 991
)
. A
position-effect
assay for
boundaries of
higher
order chromo-
somal
domains. Cell 64,941-950.
Pikaart,
M. J., Recillas
-Targa,
F., and Felsenfeld, G.
(
I 998
)
. Loss of transcriptional
activity of a
transgene is accompanied
by
DNA methyla-
tion and histone deacetylation and is
pre-
vented by insulators. Genes Dev. 12,
2852-2862.
Zhao,I(, Hart,
C.
M.,
and
Laemmli,
U. K.
(1995).
Visualization
of chromosomal domains with
boundary element-associated
factor BEAF-12.
Cell 81, 879-889.
Insutators
May Act in One Direction
Resea
rc h
Gerasimova,
T. I., Byrd, I(., and Corces, V. G.
(2000).
A chromatin
insulator determines the
nuclear localization
of DNA. Mol. Cell 6,
r025-to)5.
Harrison, D. A., Gdula,
D. A., Cyne, R. S., and
Corces,
V.
G.
(1993).
A leucine zipper domain
of the suppressor
of hairy-wing
protein
medi-
ates
its repressive effect
on enhancer function.
Genes
Dev. 7, 1966-197 8.
Roseman,
R. R., Pirlotta, V., and Geyer,
P. K.
(19931.
The su(Hw)
protein
insulates
expres-
sion of the
D melanogaster
white
gene
from
chromosomal
position-effecrs.
EMB) J. 12,
435-442.
Insulators Can
Vary in Strength
Resea
rch
Hagstrom,
I(., Muller, M., and Schedl, P.
(1996).
Fab-7 functions as a chromatin domain
boundary
to ensure
proper
segment speci{ica-
tion by the
Drosophila bithorax complex. Genes
Dev.
10,3202-)215.
Mihaly, J. et al.
\1997).
In situ drssection of the
Fab-7 region of the bithorax complex
into a
chromatin domain boundary and a
polycomb-
response element. Development 124,
l
809-l 820.
Zhou, J. and
Levine, M.
(1999).
A novel czi-
regulatory element, the PTS, mediates an anti-
insulator activity inthe Drosophila embryo.
Cell99,567-575.
DNAase Hypersensitive Sites Reflect
Changes
in
Chromatin
Structure
Review
Gross,
D. S. and Garrard, W T.
(1988).
Nuclease
hypersensitive sites in chromatin. Annu Rev.
Biochem. 57, 159-197.
Resea
rch
Groudine, M., and Weintraub, H.
(1982).
Propaga-
tion of
globin
DNAase I-hypersensitive
sites
in
absence of
factors required for induction:
a
possible
mechanism for
determination. Cell
30, t3t-t39.
Moyne, G., Harper,
F.,
Saragosti, S., and Yaniv M.
(
I 982
).
Absence of
nucleosomes
in a histone-
containing nucleoprotein complex obtained
by dissociation of
purified
SV40
virions.
Cel/
30,
r23-r30.
Scott, W.
A.
and Wigmore,
D.
J.
(1978).
Sites in
SV40 chromatin which are
preferentially
cleaved by endonucleases.
Cell
15,
l5l l-1518.
Varshavsky, A. J., Sundin, O., and Bohn, M.
J.
(1978).
SV40
viral minichromosome:
prefer-
ential exposure of the origin of replication as
probed
by restriction
endonucleases. Nucleic
Acids Res.
5, )469-)479.
794 CHAPTER 29 Nucteosomes

Domains
Define
Regions
That
Contain
Active
Genes
Research
Stalder, J. et
al.
(1980).
Tissue-specific
DNA
cleav-
age in
the
globin
chromatin
domain
intro-
duced
by DNAase
I.
Cell 20, 45t-460.
An LCR
May
Control
a Domain
Reviews
Bulger,
M. and
Groudine,
M.
(1999).
Looping
ver-
sus
linking:
toward a
model for
long-distance
gene
activation.
Genes Dev. Lj^,2465-2477.
Grosveld, F., Antoniou,
M., Berry,
M., De Boer,
E.,
Dillon,
N., Ellis,
J., Fraser, P.,
Hanscombe,
O..
Hurst,
J., and Imam,
A.
(1993).
The regulation
of human
globin gene
switching.
Philos. Trans.
R. Soc Lond.
B Biol.
Sci. )39, t8)-t9t.
Research
Gribnau,
J., de
Boer, E., Tfimborn, T.,
Wijgerde,
M.,
Milot, E.,
Grosveld, F., and
Fraser,
P.
(I998).
Chromatin interaction mechanism of tran-
scriptional
control in vitro. EMBO J. 17,
6020-6027.
Spilianakis,
C. G.,
Lalioti, M. D., Town, T., Lee,
G. R.,
and
Flavell, R. A.
(2005).
Interchromo-
somal
associations between alternatively
expressed loci.
Nature
475, 637
-645.
van Assendelft,
G. B., Hanscombe, O., Grosveld,
F.,
and
Greaves, D.
R.
(I989).
The
B-globin
domi-
nant
control region activates
homologous
and
heterologous
promoters
in
a tissue-specific
manner.
Cell 56, 969-977.
What
Constitutes a
Regulatory Domain?
Review
West, A. G., Gaszner, M., and Felsenfeld, G.
(2OO2l
.Insulators: many functions/
many
mechanisms.
Genes
Dev. 16, 27 1-288.
References 795

Controlting
Chromatin
Structure
CHAPTER OUTLINE
Introduction
Chromatin Can
Have
Atternative States
.
Chromatin structure is stable and
cannot
be changed by
altering
the equitibrium of transcription factors and
histones.
Chromatin Remodeting Is an Active Process
r
There are several
chromatin
remodeling
complexes that use
energy
provided
by
hydrotysis
of
ATP.
r
The SWI/SNF, RSC.
and
NURF
comptexes atl are
very
large,
and they share some
common subunits.
.
A remodeling
comptex does not
itself
have specificity
for
any
particular
target site, but must be recruited by a compo-
nent
of the transcription apparatus.
Nucteosome
0rganization
May Be
Changed
at the Promoter
o
Remodeting
comptexes are recrujted to
promoters
by
sequence-specjfi
c activators.
r
The factor
may be released
once the
remodeting
complex
has
bound.
r
The MMTV
promoter
requires
a change
in rotational
position-
ing of a nucteosome
to a[[ow an activator
to
bind to DNA on
the nucteosome.
Histone
Modification Is
a
Key
Event
r
Histones
are modified
by
methylation,
acetylation, and
phosphory[ation.
Histone
Acetytation
0ccurs in Two
Circumstances
o
Histone
acetytation
occurs transjentlyat replication.
o
Histone
acetylation
is associated with activation of
gene
expresslon.
Acetytases
Are Associated
with Activators
r
Deacetytated
chromatin may have
a more condensed
structu
re.
r
Transcription
activators
are associated with histone acety-
lase
activities in [arge
complexes.
o
Histone
acetylases
vary in their
target specificity.
o
Acetylation
coutd
affect transcription in a
quantitative
or
quatitative
wa;.
Deacetylases
Are Associated with Repressors
o
Deacetytation is associated with
repression
of
gene
activity.
r
Deacetylases are
present
in comptexes with
repressor
activity.
Methytation
of
Histones and
DNA Is
Connected
.
Methytation of both
DNA
and
histones is a feature of inac-
tive
chromatin.
o
The
two types
of methytation event
may
be connected.
Chromatin States
Are Interconverted by Modification
o
Acetytation of histones is associated
with
gene
activation.
.
Methylation of
DNA
and of
histones is associated with
heteroch ro mati n.
Promoter Activation
Involves
an 0rdered Series of
Events
.
The remodeting comptex
may recruit
the acetytating
complex.
.
Acetytation of histones
may
be the event that
maintains
the
complex
jn
the actjvated state.
Histo ne Phosphorylation
Affects
Ch
romati n
Structure
o
At
least two
histones are targets
for
phosphorylation, possi-
bty with opposing effects.
Some Common
Motifs Are Found in Proteins That Modifv
Chromatin
.
The chromo domain is
found in
several chromatin oroteins
that have ejther activating or
repressing
effects on
gene
expressron.
.
The SET domain is
part
of the catatytic site of
protein
methyttra nsferases.
.
The bromo domain
is found in
a
variety
of
proteins
that
interact with chromatin and
is
used to
recognize
acetylated
sites on histones.
Summary
796

Introduction
When
transcription is
treated in
terms
of
inter-
actions involving
DNA and
individual
transcrip-
tion factors
and RNA
polymerases,
we
get
an
accurate description
of the
events that
occur ilt
vitro,but this lacks
an important
feature
of tran-
scription in vivo.
The cellular
genome
is or-
ganized
as nucleosomes,
but initiation
of
transcription
generally
is
prevented
if the
pro-
moter region is
packaged
into
nucleosomes.
In
this sense, histones function
as
generalized
repressors
of transcription
(a
rather
old idea),
although
we see in this
chapter that
they are
also involved
in more
specific interactions.
Acti-
vation of a
gene
requires
changes
in the state
of
chromatin: The
essential issue
is how
the tran-
scription factors
gain
access to the
promoter
DNA.
Local
chromatin
structure is an integral
part
o{ controlling
gene
expression.
Genes may exist
in either
of two structural
conditions. Genes
are
found in
an
"active"
state only in
the cells in
which they
are expressed. The
change of struc-
ture
precedes
the act
of transcription,
and indi-
cates that the
gene
is
"transcribable."
This
suggests that acquisition
of the
"active"
struc-
ture must be the first
step in
gene
expression.
Active
genes
are
found
in
domains of euchro-
matin
with a
preferential
susceptibility
to
nucle-
ases
(see
Section
29.l9,Domains
Define Regions
That
Contain Active
Genes). Hypersensitive
sites
are created at
promoters
before
a
gene
is acti-
vated
(see
Section 29.I8, DNAase
Hypersensi-
tive Sites Change
Chromatin Structure).
More recently
it has turned
out that there
is an intimate and
continuing
connection
between initiation
of transcription
and chro-
matin
structure. Some
activators of
gene
tran-
scription directly modify
histones; in
particular,
acetylation
of
histones
is associated
with
gene
activation. Conversely,
some repressors
of
tran-
scription function
by deacetylating
histones.
Thus
a reversible change in
histone structure
in
the vicinity of the
promoter
is involved in
the control
of
gene
expression.
This
may
be
part
of the mechanism by
which a
gene
is main-
tained in
an
active or inactive state.
The mechanisms by which local
regions of
chromatin
are maintained
in an inactive
(silent)
state are related to the means
by which an indi-
vidual
promoter
is repressed. The
proteins
involved
in the formation of
heterochromatin
act on
chromatin
via the histones,
and modifi-
cations of the
histones may be an
important
feature in the interaction.
Once established,
such changes in chromatin
may
persist
through
cell
divisions, creating
an epigenetic
state in
which the
properties
of
a
gene
are determined
by the self-perpetuating structure
of chromatin.
The name
epigenetic
reflects the
fact that a
gene
may have
an
inherited condition
(it
may be
active or may be inactive)
that does
not
depend
on
its
sequence.
Yet a further
insight into epi-
genetic properties
is
given
by the self-perpetu-
ating
structures
of
prions
(proteinaceous
infectious
agents).
Chromatin
Can
Have
Alternative
States
.
Chromatin
structure
is stable
and cannot be
changed by
altering the equitibrium
of
transcription
factors and
histones.
Two types of models
have been
proposed
to
explain
how the state of
expression of
DNA
is changed: equilibrium
and
discontinuous
change-of-state.
',r
.,
shows
the equilibrium
model.
Here the
only
pertinent factor
is
the
concentra-
tion
of the
repressor or activator
protein,
which
drives an equilibrium
between
free
form and
DNA-bound form. When
the concentration
of
the
protein
is high enough,
its DNA-binding
site
is occupied, and the
state of
expression of
the
In an equitibrium model.
the state ofa binding site on
DNA depends
on the con-
centratjon of
the
protein
that bjnds to it.
30.2
Chromatin
Can
Have Alternative
States
797
DNA
is affected.
(Binding
might either repress
or activate any
particular
target sequence.) This
type of model explains the regulation
of
tran-
scription in bacterial cells, where
gene
expres-
sion is determined exclusively by the actions of
individual repressor and
activator
proteins (see
Chapter I2, The
Operon). Whether a bacterial
gene
is
transcribed can be
predicted
from the
sum of the
concentrations of the
various
fac-
tors that either activate or repress the individ-
ual
gene.
Changes in these
concentrations a/
any time will
change the state of expression
accordingly.
In most cases, the
protein
binding
is
cooperative, so that once
the concentration
becomes high
enough, there is a rapid associa-
tion with DNA, resulting in
a switch
in
gene
expressron.
A
different situation applies with eukary-
otic
chromatin. Early in vitro
experiments
showed that either an
active or
inactive
state
can
be established, but this is not
affected by
the subsequent addition
of other components.
The transcription
factor TFnyA,
which
is required
for RNA
polymerase
III to transcribe 5S rRNA
genes,
cannot activate its target
genes
in
vitro
if,
they are complexed
with
histones.
If the factor
is
presented
with free DNA, though, it forms
a
transcription complex,
and then the addition
of histones
does
not
prevent
the
gene
from
remaining
active.
Once the factor has
bound,
it remains
at the site;
this allows
a succession of
RNA
polymerase
molecules to initiate transcrip-
tion. Whether the
factor
or
histones
get
to the
control site first may be the critical factor.
l:*{.!ft1, .l,J.J
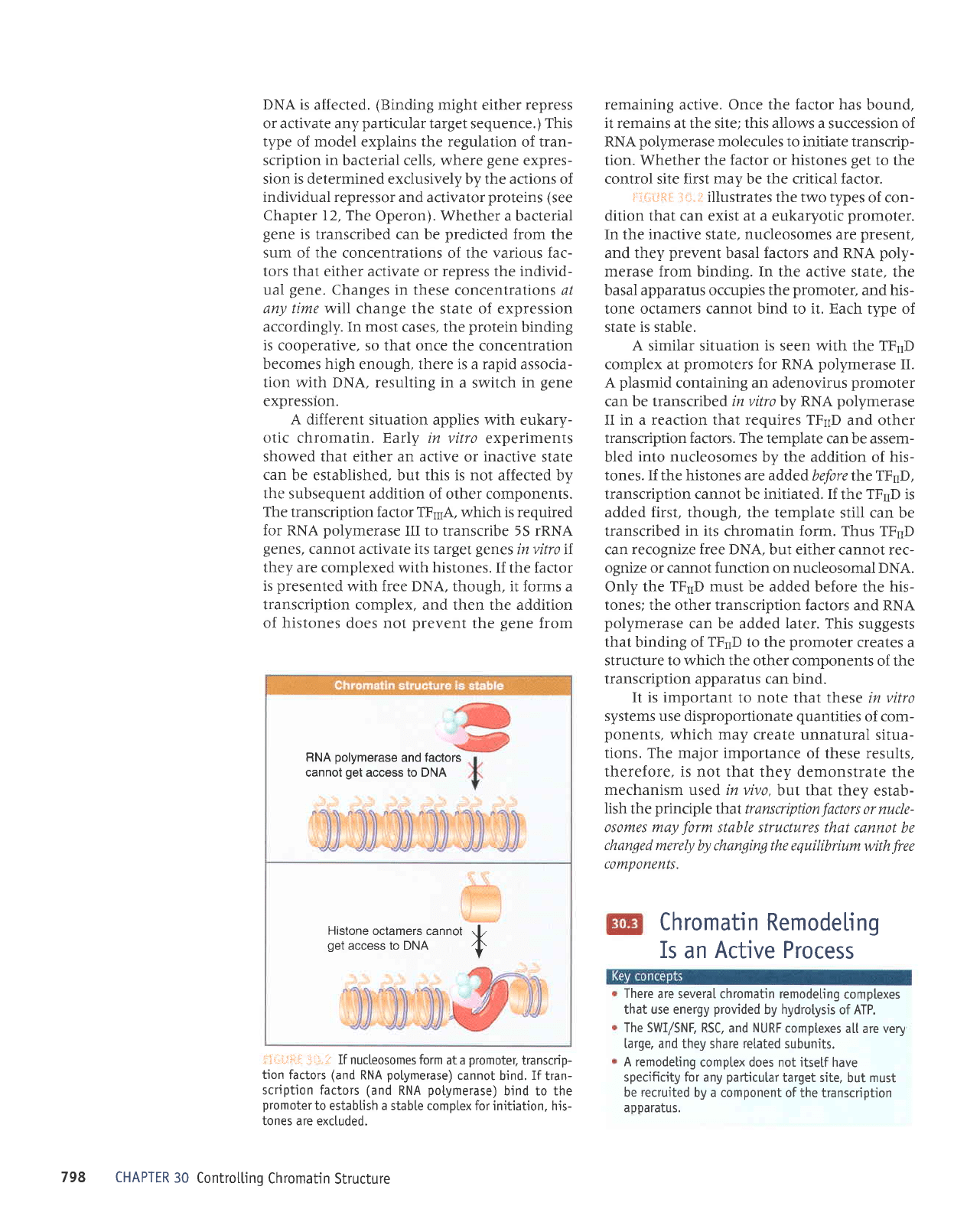
illustrates the two types
of con-
dition that can exist
at a
eukaryotic
promoter.
In the inactive state, nucleosomes are
present,
and they
prevent
basal
factors
and RNA
poly-
merase from binding. In the active
state. the
basal apparatus occupies
the
promoter,
and his-
tone octamers cannot bind to it. Each tvne of
state is stable.
A
similar situation
is seen
with the TF1D
complex at
promoters
for RNA
polymerase
II.
A
plasmid
containing an adenovirus
promoter
can be transcribed in vitro by RNA
polymerase
II in a reaction that requires TF11D
and other
transcription factors. The
template can be assem-
bled
into
nucleosomes by the addition
of
his-
tones. If
the
histones are added
before tl:'eTF1D,
transcription cannot be initiated. If
the
TF1D
is
added first, though, the
template still can be
transcribed
in its
chromatin form. Thus TF11D
can recognize free DNA, but either
cannot
rec-
ognize or cannot function on nucleosomal
DNA.
Only the TFnD must be added before
the his-
tones; the other transcription factors
and RNA
polymerase
can be added
later.
This suggests
that
binding of
TFnD
to the
promoter
creates
a
structure to which the other components
of the
transcription apparatus can bind.
It is important
to note that these
in
vitro
systems use disproportionate
quantities
of com-
ponents,
which
may
create unnatural
situa-
tions. The major importance
of these results,
therefore, is not that they
demonstrate the
mechanism used iz vivo,bul
that they
estab-
lish
the
principle
Lhat transcription
factors
or nucle-
osomes may
form
stable structures
that cannot be
ch
ang e d mer e ly by ch ang ing the e
q
ui lib rium
with
fr
e e
cjmpjnents.
Chromatin RemodeLing
Is
an
Active Process
There
are several chromatin remodeling
complexes
that use energy
provided
by
hydrotysis
of ATP.
The SWI/SNF,
RSC, and
NURF
comptexes
a[[ are very
[arge, and
they share retated subunits.
A remodeling
comptex does not itsetf have
specificity for any
particutar
target site,
but
must
be
recruited
by a component
of the transcription
appararus.
RNA
polymerase
and factors
cannot
get
access to DNA
Histone
octamers cannot
,f,
get
access to DNA
0
,r!i:.iiitii
Jil..i
If nucteosomes form
at a
promoter.
transcrip-
tion factors
(and
RNA
potymerase)
cannot
bind.
If
tran-
;cription factors (and
RNA
potymerase)
bind to the
promoter
to establish a stabte
comptex for initiation, his-
tones are excluded.
CHAPTER
30
Controtting
Chromatin
Structure
798
The
general process
of inducing
changes in chro-
matin structure is
called chromatin
remod-
eling. This consists
of mechanisms
for displacing
histones
that depend
on the input
of energy.
Many
protein-protein
and
protein-DNA
con-
tacts need
to be disrupted
to release histones
from chromatin. There
is no free
ride: The
energy must be
provided
to disrupt
these con-
tacts.
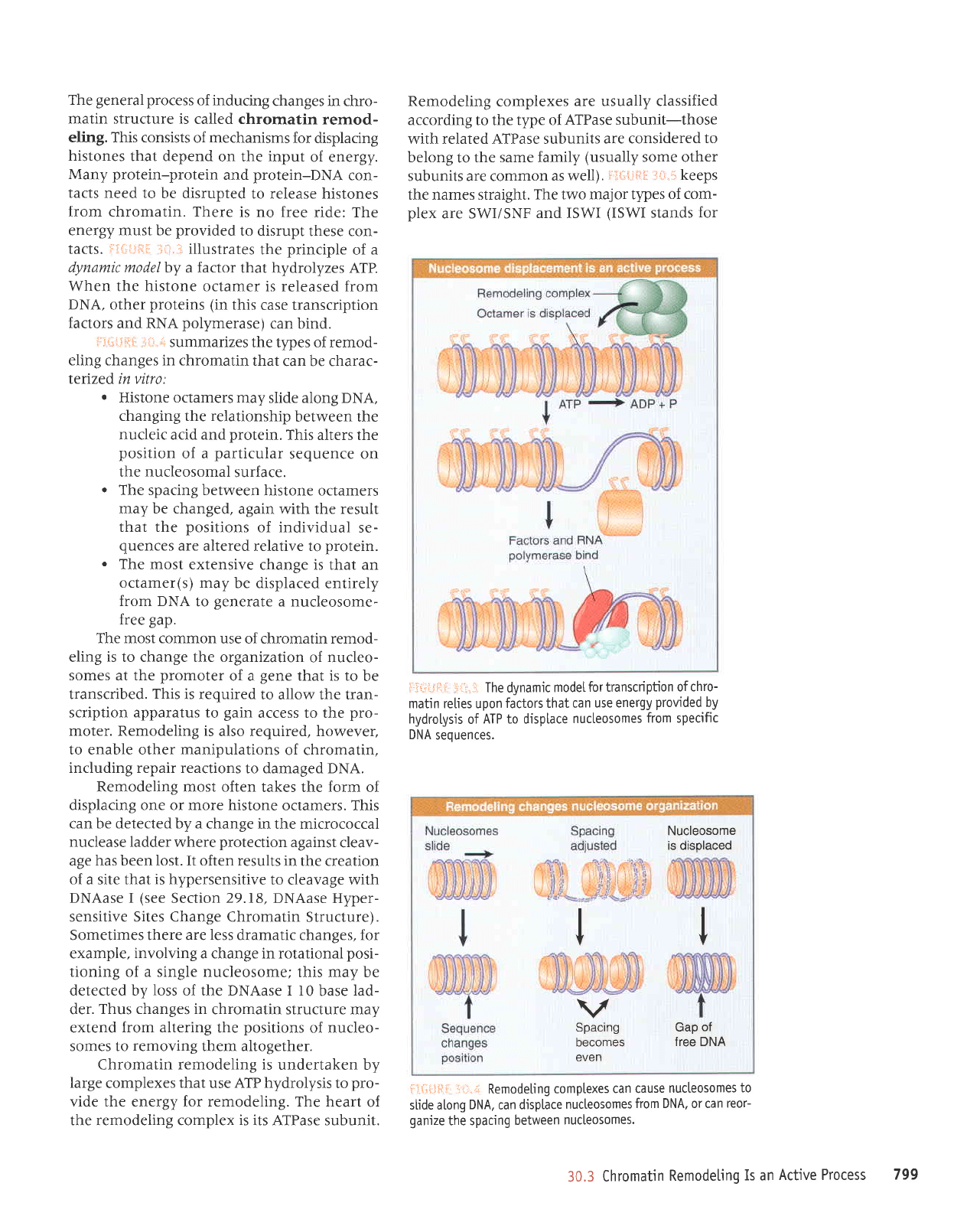
fii-:i":{il
.:!{.i,::l
illustrates
the
principle
of
a
dynamic
model by a factor
that hydrolyzes ATP.
When the histone
octamer is released from
DNA,
other
proteins (in
this
case transcription
factors
and RNA
polymerase)
can bind.
i"5i;i,Ji'ti
.:1,.ii..:.'
51m-".izes
the types of remod-
eling changes in chromatin
that
can be charac-
terized
in
vitro:
.
Histone octamers
may slide along DNA,
changing the relationship
between the
nucleic acid and
protein.
This alters the
position
of a
particular
sequence on
the nucleosomal
surface.
.
The spacing between
histone octamers
may be changed,
again with the result
that the
positions
of individual
se-
quences
are
altered relative to
protein.
.
The most extensive
change is that an
octamer(s) may
be displaced
entirely
from DNA
to
senerate
a nucleosome-
free
gap.
The most common
use of chromatin remod-
eling
is
to change the organization
of nucleo-
somes at the
promoter
of a
gene
that is to
be
transcribed.
This
is required
to allow the tran-
scription apparatus to
gain
access to the
pro-
moter. Remodeling is
also required, however,
to enable other manipulations
of chromatin,
including repair reactions
to damaged DNA.
Remodeling most often
takes the
form
of
displacing one or more histone
octamers. This
can be detected by a
change in the micrococcal
nuclease ladder
where
protection
against
cleav-
age has been lost. It often results
in the creation
of a site that
is
hypersensitive
to cleavage with
DNAase I
(see
Section
29.I8,
DNAase Hyper-
sensitive Sites Change Chromatin
Structure).
Sometimes there are
less
dramatic changes, for
example,
involving
a change in rotational
posi-
tioning of a single nucleosome;
this may be
detected by
loss
of the DNAase I I0
base
lad-
der.
Thus changes in
chromatin structure may
extend
from altering
the
positions
of nucleo-
somes to
removing
them altogether.
Chromatin
remodeling is
undertaken by
large
complexes
that
use
ATP
hydrolysis to
pro-
vide the energy for remodeling. The heart
of
the remodeling complex
is
its ATPase subunit.
Remodeling
complexes
are usually classified
according
to the
type of ATPase
subunit-those
with related ATPase subunits
are considered
to
belong
to
the same family
(usually
some other
subunits are common
as well).
| lillii]$
::l+.ii
keeps
the names straight.
The two major tlpes
of com-
plex
are SWI/SNF
and ISWI
(ISWI
stands
for
ii{.:i.:iii
:ii.'!.-i The dynamic
model
for transcription
of chro-
matin reties upon
factors that can
use energy
provided
by
hydrotysis
of
ATP to disp[ace
nucteosomes
from specific
DNA
seouences.
+'iiltJitF
:!Li.+ Remodeling
comptexes
can cause
nucleosomes
to
slide along
DNA,
can
displace
nucleosomes
from DNA. or can
reor-
ganize
the spacing
between
nucleosomes.
Nucleosome
is displaced
i
I
T
I
v
Gap
of
free DNA
,r.
r
,i,'\
'i
!l
rl.;
I
I
v
V
Spacing
becomes
30.3
Chromatin
Remodelinq
Is an
Active Process
799
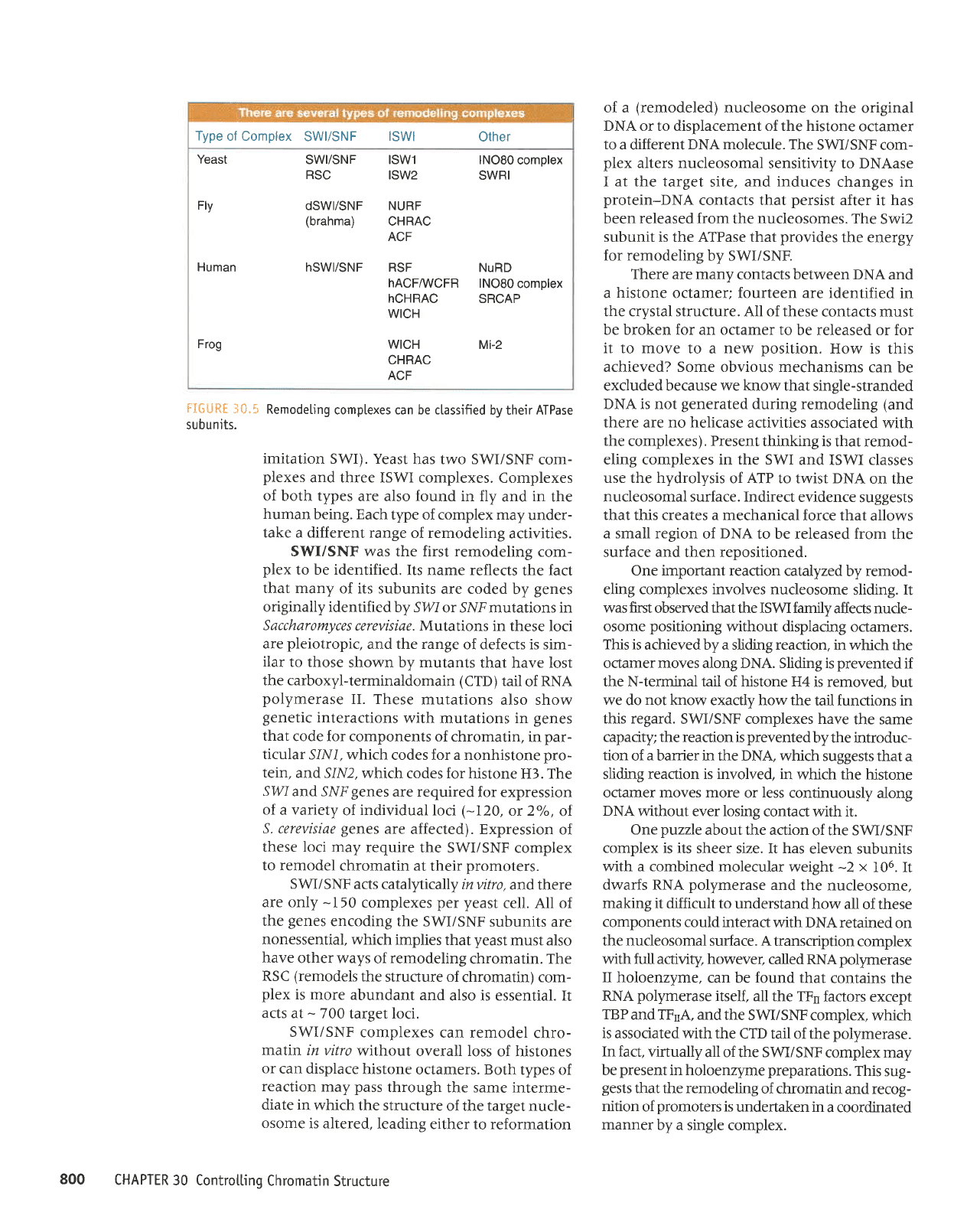
Type
of Complex
SWI/SNF
lSWl
Other
Yeast
SWYSNF lSWl lNO80
complex
RSC
ISW2
SWRI
Fly
dSW|/SNF NURF
(brahma)
CHRAC
ACF
Human
hSW|/SNF RSF
NuRD
hACFAIVCFR
lNO80
complex
hCHRAC
SRCAP
WICH
Frog
WICH
Mi-2
CHRAC
ACF
tIGiJRE
ii0.5
subunits.
Remodeling
complexes can be classified
by their ATPase
imitation
SWI). Yeast
has two SWI/SNF
com-
plexes
and three ISWI
complexes. Complexes
of both types
are also
found
in fly
and
in
the
human
being. Each
type of complex may
under-
take a different range
of remodeling
activities.
SWI/SNF
was
the first remodeling
com-
plex
to be identified.
Its name reflects
the
fact
that many
of its subunits
are coded
by
genes
originally identified
by SWI
or SNF mutations in
Saccharomyces
cerevisiae. Mutations
in these loci
are
pleiotropic,
and the range
of defects is sim-
ilar
to those
shown by mutants
that have lost
the carboxyl-terminaldomain (CTD)
tail
of
RNA
polymerase
II. These
mutations
also show
genetic
interactions
with mutations in
genes
that
code for components
of chromatin,
in
par-
ticular
SINI, which
codes for a nonhistone
pro-
tein,
and SIN2,
which codes for
histone H3. The
SI4'1
and SNF
genes
are required
for expression
of
a variety
of
individual
loci
(-120,
or 2"h,
of.
S. cerevisiae
genes
are
affected). Expression
of
these loci
may require
the SWI/SNF
complex
to remodel
chromatin at
their
promoters.
SWI/SNF
acts catalytically
invitro,
and there
are only
-150
complexes
per
yeast
cell. All of
the
genes
encoding the
SWI/SNF
subunits are
nonessential, which
implies
that
yeast
must aiso
have
other ways
of
remodeling
chromatin. The
RSC
(remodels
the
structure of
chromatin) com-
plex
is
more abundant
and
also is essential. It
acts
at
-
700
target loci.
SWI/SNF
complexes
can remodel
chro-
matin
in vitro
without
overall loss
of histones
or
can displace
histone
octamers. Both
types of
reaction
may
pass
through
the same interme-
diate in
which
the structure
of the target nucie-
osome
is altered,
leading
either to reformation
of a
(remodeled)
nucleosome
on the original
DNA or to displacement of the histone
octamer
to
a different
DNA molecule.
The S\M/SNF
com-
plex
alters nucleosomal
sensitivity to DNAase
I at the target
site, and
induces
changes in
protein-DNA
contacts that
persist
after it
has
been
released
from the nucleosomes. The
Swi2
subunit is the ATPase that
provides
the energy
for remodeling
by SWI/SNF.
There are many
contacts between DNA
and
a histone octamer; fourteen are identified
in
the crystal structure. All of these
contacts must
be broken for an octamer to
be released or for
it to move
to a
new
position.
How is
this
achieved? Some obvious mechanisms
can
be
excluded because we know that
single-stranded
DNA is not
generated
during remodeling (and
there
are
no helicase
activities associated
with
the
complexes). Present thinking is
that remod-
eling complexes in the SWI
and ISWI classes
use the hydrolysis
of ATP to twist DNA
on the
nucleosomal surface. Indirect
evidence
suggests
that this creates a mechanical
force that
allows
a small region
of
DNA
to be released from
the
surface and then repositioned.
One important reaction
catalyzed
by
remod-
eling complexes involves nucleosome
sliding.
It
was first
observed that the IS\M family
affects nude-
osome
positioning
without
displacing octamers.
This is
achieved by a sliding reaction, in which
the
octamer moves along DNA. Sliding
is
prevented
if
the N-terminal tail of histone
H4 is removed,
but
we do
not
know exactly how the
tail functions in
this regard.
SWI/SNF complexes have
the same
capacity; the reaction
is
prevented
by the introduc-
tion
of
a
barrier
in
the DNA" which
suggests that
a
sliding reaction is involved, in
which the
histone
octamer moves more
or
less
continuously
along
DNA
without ever losing
contact with it.
One
puzzle
about the action
of the S\M/SNF
complex is its
sheer size. It has
eleven subunits
with a combined molecular
weight
-2
x
106. It
dwarfs RNA
polymerase
and
the nucleosome,
making
it difficult to
understand how
all of these
components could interact
with DNA retained
on
the nucleosomal
surface. A transcription
complex
with
full
activity, howeve(,
called RNA
polymerase
II
holoenzyme,
can be found that
contains
the
RNA
polymerase
itself, all the TFn
factors
except
TBP
and TFnA, and
the S\M/SNF
complex,
which
is
associated with the
CTD tail of the
polymerase.
In
fact, virtually
all of the S\M/SNF
complex
may
be
present
in holoenzyrne
preparations.
This
sug-
gests
*rat the remodeling
of chromatin
and recog-
nition
of
promoters
is
undertaken in
a coordinated
manner
by a
single complex.
800
CHAPTER
30 Controtting
Chromatin
Structure
