Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.


Ig9
surputo6
6urpurg-ypq;o
sadAt
fue4
ary
olaql
g.9Z
'lFnqns
unf eql
Surreld:oqdsoqd
z(q
ruroJ
J^rtJe eql 01
pJlJeluo)
sr
(sog pue
unf
slrunqns
aql
uJJMlJq
rJurporJlJq
p)
IdV
'uorte1,{
-roqdsoqd,{q
ruroy
alr]lp
eq1 ot
pJuJA
-uo)
sr
JJSH
'uorleJ$lpotu
,{q
pallorruot
,(lnarrp
aq deru
JoDeJ e
Jo
,hmrDe egJ .
'suratord
ureruop
-oeuoq
sE r{Jns
'lueurdola,Lep
Jtel
-n8ar
1eql
srope;;o pld,{t
sr
srql
'[e)
Jo
ad.{t relnrqred
e ur Lpo
pazrsaqtuLs
sr
lr
JSne)Jq
rryrtads-anssrl
sr rolf,e]
11
o
:eT'bi
3HC]:;I+
ur .{.lletrlpruJqts pJlprl
-snllr
se
's.deu
leranas
Jo
euo.due
ur
paleln8ar
aq
,{.eru
role,LtlJe
Jlqr)npur
ue
;o
,{.1rnrDe
aq1
'yNC
01 Surpurq
ur
penlolur
sr
teql
sJnprsJJ pa8reqr
Llaarlrsod
Jo
q)laJls
e sr raddrz qJeJ
01
luarefpy
'Jerurp
e
ruroJ
01 aprrdadz(lod
Jeqloup
ur raddrz B
Q1I1V\
sDeralut
aprldaddlod
auo ur rad
-drz
aunnal
y
'uorlrsod
qlueAJS
^re^e
ur
enprsal
eurf,nJl
p
r{lrM sprJe
oullue
Jo
qJtarts
p
Jo
tsrsuoJ
sraddlz eulJnal o
.VNC
slJpluoJ
Jrloru
slql reau
uor8ar Jrseq
p
pup
'ezrJerurp
ol surJloJd
salqeua;r1oru
JqJ
'spDe
ourrup
gz
01
zI
tuoJJ serJp^
dool
SurpauuoJ
aqt
Jo
q18uJl
JqJ
'Jprs
Jaqlo
Jqt
uo senprseJ
pa8reqr
pue
eprs
euo
uo sanprsJJ
lrqoqdorp,{q
Jo
JJeJ e
sluasard
xrTJrI
Jrqtedrqdue qJp[I
'surel
-ord
Surpurq-ygq
rrlofualne
roJ Surpor
saua8
ur
pue
sroleln8ar
leluarudolarrap
eruos ur
pJrJuuJpr
uaaq seq
Jlrour
(gfg)
xrlaq-doo1-xllaq
rrqtedrqdrup
er{l o
'sJolJPJ
UOIIOrJJS
-ueJl
uPrleruluPru
JoJ sauaS
ur
luasard
oslp sr
11
'ultt[dosotO
ur uorteln8ar
1e]
-uarudolanap
qtp,r peuJJf,uo)
saua8
,(q
pepo)
suralord
IpJJAJs
ur
pezrrJpeJpqJ
lsrr;
aruanbas
p
'urpluopoatuoq
aql
ur
luasard
sr
Jrlotu
aql
Jo
tuJoJ
paleleJ
V
'vNO
ssoJf,p a13ue
ue
le
sJrl Jer{lo
eql lVN(
;o
anoor8
roferu
eq1 ur sJrl
xrTJq-n
aug
'srossardar
a8eqd;o urpuop
Sutpurq-yg(
eql
sp
prrJrturpr
L11eu
-r8tro
se,vr
Jlroru
xllaq-urnl-xlaq
e{I
.
'pua64
ilaws
g
spqq
U Urun
a^tpaw e fij
-nt
utalotd at11
:ryuatadl
snplLu
leraua8
eurps
Jqt
qll,lr
srotelrDe pJte,LurB-puB8rl
yo
,{puey,radns
aql
Jo
sJJqrueu
aJe sJol
-da:al
proJets
aql rordatar
prf
p
JtoutlJJ
aq1 Jo rolde)eJ
JuoruJoq pro.rLqr
aqr
sp
qJns
'sroldarar
Jeqlo
qlrM
raqlaSol
'paz^,{.1eue
.d11ny
lsoru
eql sr rotdarar
pIoJruoJof,nlS
aql'proJJls
relnlrlred
e Supurq,{q
pale,Lrpe
sr
rotdarar
qreg
:drqsuorlelar
lpuorlJunJ
p
^q dnorE e
sp
paurJep
are sroldalar
plorels
ar{J .
's.roldarar
prorels
eql
q
os1e
punoJ
sr
Jrtou
aql
Jo
ruJoJ
lJullsrp
y
'(sror
-rey
uorldursueJl
paunsJrd pue)
sJopeJ
uorldrrlsuerl
Jaqlo
IprJnJS
ur
perJrluapr
ueaq eJurs seq
1I
'saua3
yNUJ
SE
eqrJJs
-ueJl
01
m
aseraur.dlod
y51g
ro;
pa;mbar
sr
qJrqm
'yurg1
JolJeJ ur
pazruSo
-rar
,{.1pur8uo sem
lI
'ureruop
Surpurq
-VN11
e sasrrdruor;ltoru,ra8q;
)ufz
Jr{l
r
:yNCI
ol Eurpurq ro; alqrsuodsar
sr
tnpruop sql ul
Jlloru
uoqs,{larrBelar
e
'praua8
u1
'ureuop
Surpurq-ygq
yo
addl aql
o1 Surprone
parJrsselJ
ualJo JJp srollpd
'uolldrnsuerl
3ur
-tp^ltJp
JoJ
pup
vN11
01 Supurq ro; alqrsuodsar
JJP sureuop
luerJJJrp
qrlq^
ur arnl)nJls rel
-npou p
e^pq 01 JolpnrlJp ue JoJ uoruluo)
sl
1I
'sa1rs
1e6re1
lpnphrpur
ro; r\logoads
laJuol
leql
Jqou
lgpaos e
Jo
suorleuP^
aruanbas aneq dnor6 aues eqlJo sraQua[rrl
o
'utPur0p
Durpurq-vN0
to
adfl aq1 o1 burprorre
pagrsspll
orp sro]pAr.]l!
o
sur.Pruo0 6urpurg-vNo
Jo
sadAl
Aue6
alv araql
'xeloruoJ
uorlelllul ar{l
Jo
uou
-pruroJ
lsrsse
JsrMJJqlo Jo azrTrqels
lpq]
suoll
-JeJalur
uralord-uralord
;o
enurl
,{q
z(lqeqord
'snleredde
uortdrrlsuerl
pseq
aqt.dq uouelqul
;o,{ruang;a
aql aspaJJur o1
z(lluapuadapur
JIqe
sr
lueruJlJ
auo z(ue o1 Surpurq JolJeJ
p
teqt rsa8
-8ns
stuaura8uerre alrlplal Jraql
Jo
,{11gqua1y
pel[uHun
,{lluaredde
Jql
pue
'uorlJp
Jo
e]uep
-uadapur
Jreql
'sluaruala
Jo.{larren
JqJ
'sJporu
JJqto ur uorle^rl)e
paJJe
lou
sJop
uorle^rlJP
Jo
Jporu auo JoJ
papeeu
lu3ruel3
uP
Jo
eJuJsqe
aq1
'aua6
a4t aw^q)a
Qluapuadaput
uw
'rarcut
-otd
n ncuvt4ua
uv
JaLflta
w
pap)ol
's1uau,ta1a
gatat
-1ry
ptatas
to
auo [ua
leqt
aldourrd
leraua8
aq1
salpJlsnru ureuolqloflpleru
;o
uopeln8ar aq;
'sprorJls
o1 asuodsar aql
JoJ
papaeu
dlarnlosqe sr
lr lnq
'suor
p1aru,{.q peJnpur
Ia^JI
Jq} ro uorsserdxa;o
Ie^el Ipspq
Jql
DeJJp lou
seop uor8ar sq1
Jo
uorl
-ale(
'raJuequJ
ue se selpqJq
qJlq^,lt
'luroduets
eqt
lo
upansdn dq
0EZ
pelerol
EUD
e,{q
paura
-ao3
sr sJuoruJoq
proJJls
ol asuods:r aq1
'suor
plaru
Jo
aruasard
aql o1 asuodsal ur
IUW
aql ol spulq
ISJW
roDeJ JqJ
'sluJruela
aldtr
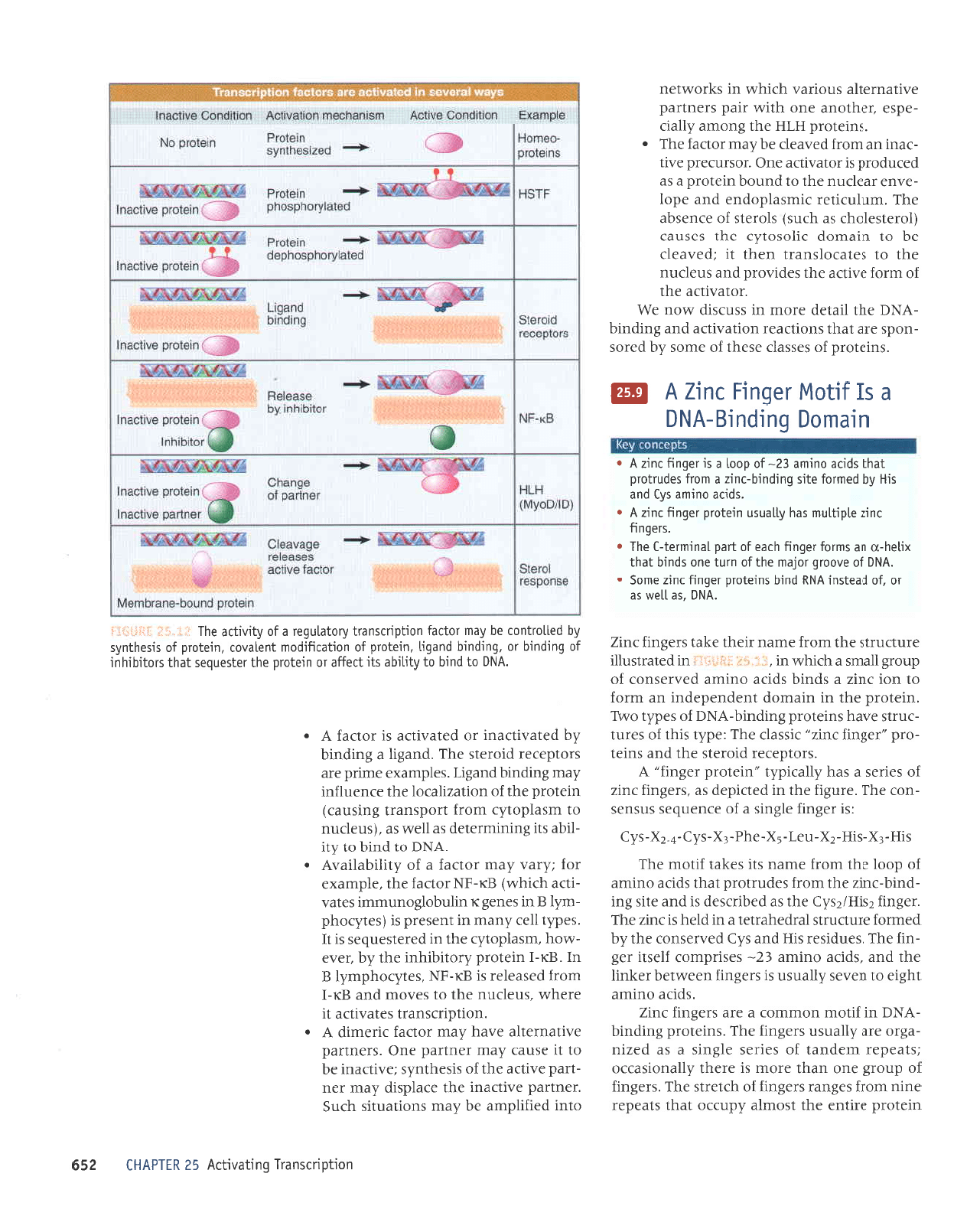
uletoJd
JrrtuJ Jqt
tsorule
.,i.dnrro
teqt
sleadar
euru ruoJJ sa8uer sra8ury
Jo
qJleJls
aq1
'sra8url
;o
dnor8
auo ueql eroru sr araql .dleuorsef,Jo
lsleadar
urepuel
Jo
serres a13urs
e se
pazru
-e8ro
are
.d11ensn
s.ra8ur; JqI
'suretord
Surpurq
-VNC
ul
Jlloru
uourtuof
p
JJp s.ra3ur.; :u17
'sprJe
oullue
tq31a
or uanas
z(11ensn
sr sra8ur; ueJMtJq
Ja>luII
aql
pue
'sprJe
ourrue
g7-
sasrrdruor
;1aslr
ra3
-ul;
eql'senprsJr srH
pup
s.d3
pan:asuor
aql
[q
peruroJ
eJnpnrls
IPJpJqerlJl
e ur
pleq
sr Jurz eqI
'ra3ur;
zsIH/zsf,l
Jqt
sp
paqrJJSJp
sr
pue
atrs 3ur
-purq-Jurz
aql uoJJ sapnlord
lpqt
splJp ounue
;o
dool aql uoJJ arueu str sJ>lpt
Jrtou
JqI
srH-€X-srH-zX-nJf
-(X-
Jq4-ty-s,{3-r-zX-s^)
:sr
ra8url a18urs e
1o
aruanbas snsuas
-uol
aqJ
'arn8r;
aqt ur
panrdap
se
'sra8ur;
rurz
Jo
selJas e seq.{11errdr(t
,,ulatord
ra8ur;.,
y
'sroldarar
proJJls
Jql
puP
surJl
-ord
,,:a8ur;
Jurz,, f,rsselJ aq1
:adz(r
srqt
}o
seJnl
-)nrts
eneq suralord Surpurq-ygq;o sad.d1 ozr41
'ulalord
Jqt ur ureuop
tuapuadapur
ue ruJoJ
01 uor )urz
e spurq
spDP ourlue
pe^rJsuo)
Io
dnoJ8
lerus
P
qJItIAl.
ur
':
i
'
:,.,
::.:;,
:-:]:,.
uI
pelPJlsnIII
arnlrnrls
Jql uorJ erueu rrJqt
a>1el sraSur;
rurT
'vN0 'sP
llaM
sP
ro
'Jo
ppelsut
VNU
pulq
suralord labug rutz ourog
r
'vNc
jo
a,roor6 roleu aq]
Jo
ulnl ouo spurq
leqt
xrloq-n up suuoJ rabug
qrea
1o
1ed
leuLLu.ral-l
aLll
r
'sleDu4
ourz a1dr11nu seq
r\11ensn
uralold re6ug lurz
V
.
'spDP
ourruP s^l
puP
srg
r\q
pauro;
alrs 6uLpurq-rurz
p
ulor1
sepnrlord
lPr]t
spllP ourulP
Ez-;o
dool e sL ta6ug )urZ
!
r
e sI
Jrlow
raburl
rulz
v @
'suratord
Jo
sJSSell
Jsaql
Jo
aruos Lq
paros
-uods
Jre
lpql
suorl)eeJ uorle^rlJe
pue
Surpurq
-Y.NO
eql
Irelap
eJoru
ur ssnJsrp 1!tou eM
'JolPArlJe
Jql
Jo
ruJoJ a^IlJe eql saptnord
pue
snal)nu
aq1 01 sJle)olsuPJl uJql
u
lps^eel)
Jq ol ulPurop:11osot,{)
Jql sJSnPJ
(loratsaloqr
se
qrns)
sloJJls
Jo
e)uesqe
JqJ
'runln)ltar
rnuseldopua
pue
adol
-J^uJ
JeJlJnu Jqt 01
punoq
uralo;d e sp
parnpord
sr Jolpnr1re
auo
'JosJnJJrd
elrl
-Jpur
ue uoJJ
pa^eJlr
aq {eu
rolJp} eef o
'suratord
H'IH
eqt Euorue .d11eo
-adsa
?aqloue auo
qt1,u
rred sraulred
eArlPUJJlle SnOrJeA
qJIqM
ur
S>IJOMIJU
uoqdursuerl
6uqeng:y
EZ
UlIdVHl Z9g
otur
paryr1drue
aq
Leu suollenlls
qJnS
'rauued
elrJJpur aql areldsrp
,{.eru rau
-ued
Jnrlle erll
Jo
slsJqlu^s
lJAItf,puI eq
o1
1r
asneJ ^,{.eru rauued
Juo
'sJeuued
JlrlpurJlle
a.Leq Leru JolJeJ
)IJerulp
V
.
'uorldrJJsuPrl
sJleArlJe
1l
aJaqM
'snJIJnu
eql o1
sJAoIu
pue
g)-I
ruoJ}
pJSPJIJJ
sr
g)r-{N
'sal{Joqdru,{l
g
uI
'g)-I
uralord
zholtqtqut aql
Iq TJ^r
-uoq
'ruseldoilr
aql ur
paratsanbas
st
11
'sadL1
lar.dueru
ur
tuasard
st
(sarrboqd
-urz(1
g
ur
saua8
y
ullnqolSounrurul
selpl
-1ne
qilq,tzr)
g)r-dN
rope} aql
'aldruexa
ro;
lLre,L
leru rotreJ
e
Jo
^llllqellpA17
o
'VNC
01
purq
ot
,{tr
-[qe
stl Suruturalap
sp
IIaM
se
'(snapnu
ot rusBldoldf, uoJJ
lrodsuerl
Sutsner)
utalord Jqt
Jo
uollpzlleJol eqt
JJuenlJuI
Leru Surpurq
pueSq
'saldruexa
atutrd
are
srotdJrJr
plorats
aq1
'pue8u
e Surputq
^dq
pale.a.rpeur
Jo
pelplrlJe
sr JolJpj
g
o
'VN6
01
pulq
ol fi1qtqe s1t
lla#e
to utelotd aq1 talsanbas
leql
sloltqtqut
1o
6uLpuLq ro
'6urpurq
puebq
'uralord
Jo
uoqelgtpoul
lualP^ol
'uLalotd
1o
stsaqlufs
r\q
pallorluor
aq fieu ro1re1
uoLldursuell
firo1e1n6at e
1o
[1ntpe aq1
i] i.'ilir
i::tr'i1jj:r
ur.Pruo0
6urpulB-vNo
(as
in
TFxyA) to
providing
just
one small
domain
consisting
of two fingers
(as
in the Drosophila
regulator
ADRI
).
The
activator
Sp I has
a DNA-
binding domain that
consists
of three zinc
fingers.
The
crystal structure
of DNA
bound by a
protein
with three fingers
suggests the struc-
ture illustrated
schematically
in
FIi:il-{&L
i.l:r.i4.
The C-terminal
part
of each
finger forms
s
helices that
bind
DNA;
the
N-terminal
part
forms
a
B
sheet.
(For
simplicity.
the
p
sheet and the
location
of the zinc ion
are not
shown in the
lower
part
of the figure.) The
three
a-helical
stretches fit into
one turn of the
major
groove;
each
o
helix
(and
thus
each finger) makes
two
sequence-specific
contacts
with DNA
(indicated
by the arrows). We
expect that
the noncon-
served amino
acids in the
C-terminal side of
each finger
are responsible
for recognizing
spe-
cific target sites.
I(nowing that zinc
fingers
are found in
authentic activators
that assist
both RNA
poly-
merases II
and
III,
we may view
finger
proteins
from
the reverse
perspective.
When
a
protein
is found
to
have
multiple zinc
fingers, there is
at least a
prima
facie
case for investigating
a
pos-
sible
role
as a transcription
factor. This
type of
identification suggests
that several
loci involved
in embryonic
development
of D. melanogaster
are regulators
oI transcription.
It is necessary
to be cautious
about interpret-
ing
the
presence
of
(putative)
zinc fingers,
though, especially
when the
protein
contains
only a single finger
motif. Fingers
may be
involved in
binding RNA rather
than DNA,
or
may not even be connected
with any nucleic
acid
binding activity. For
example, the
proto-
type zinc finger
protein,
TF11A,
binds both to
the 55
gene
and to the
product,
55 rRNA. A
translation initiation factor,
eIF2B, has a zinc
finger, and mutations
in the finger influence
the recognition
of initiation
codons. Retroviral
capsid
proteins
have
a motif related
to the
fin-
ger
that may be involved in
binding the viral
RNA.
Steroid Receptors
Are
Activators
Steroid receptors are
examples of ligand-
responsive
activators that are activated
by
binding a steroid
(or
other retated molecutes).
There are separate DNA-binding
and tigand-binding
domai
ns,
t,
\.i
@
*iJ.#
Zn+* Zn++
Zn**
Cvs
His
tl
Phe
,J
Leu
f:f*l,i*t* ;:i": ;i
Transcription
factor
5PL
has a series of
three zinc fingers,
each
with a characteristic
pattern
of
cysteine and
hjstidine residues that constitute
the zinc-
bindino site.
ftfi#Fi$ li.1!,
:1q.
Zinc
fingers may
form
cr
helices that
inseft
into
the major
groove,
which
is
assocjated
with
B
sheets
on the other side.
Steroid hormones are
synthesized
in response
to a variety of
neuroendocrine
activities and
exert major effects
on
growth,
tissue
develop-
ment, and body
homeostasis
in the
animal
world. The major
groups
of
steroids
and some
other compounds
with
related
(molecular)
activ-
ities are classified
in Ftr..iii[tI
ili.tfi.
25.10 Steroid
Receptors
Are
Activators 653
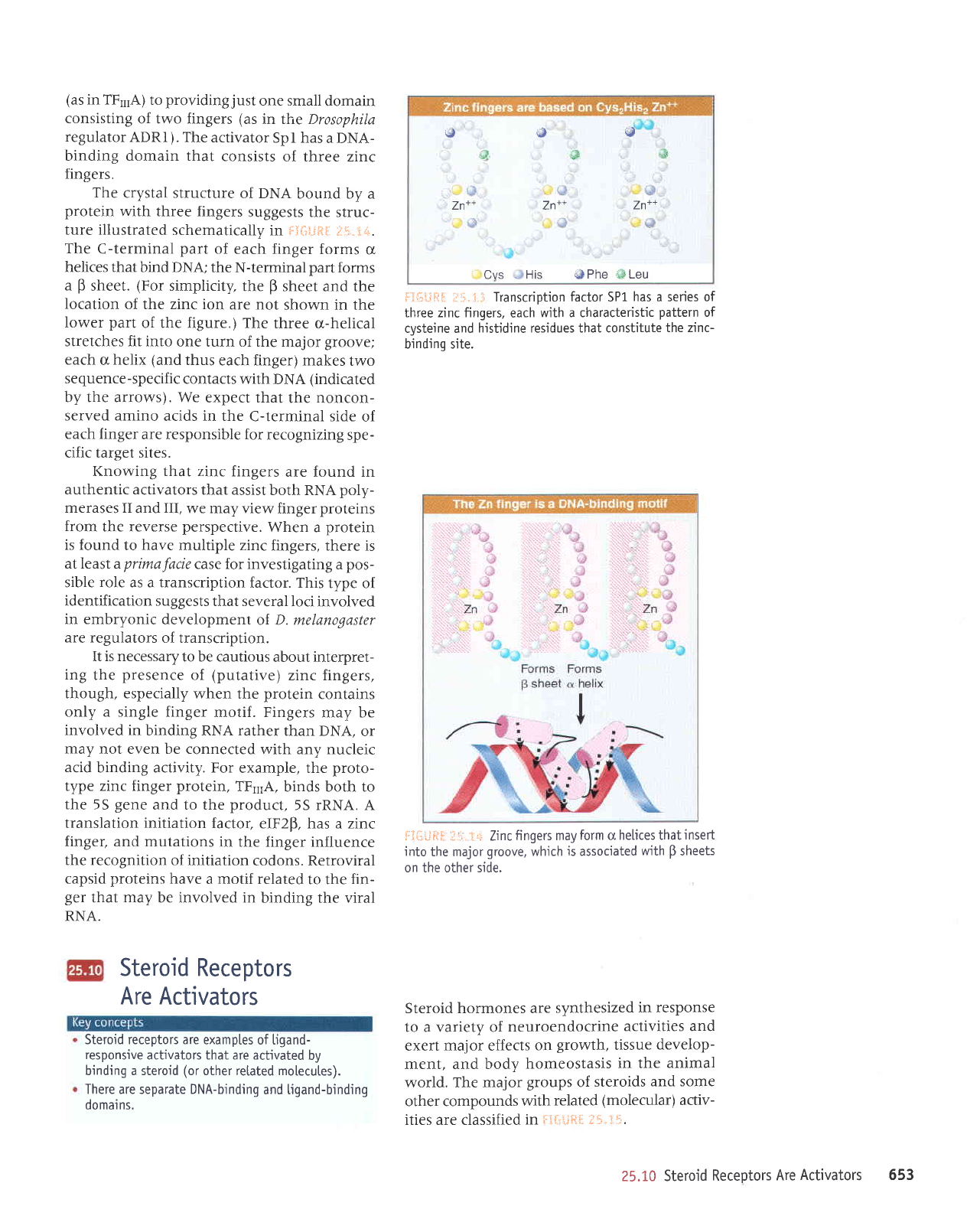
Glucocorticoids increase
blood sugar; also have
CH,OH
anti-inflammatory
action
|
-
Estrogens
are
involved
in female
sex
develooment
Androgens are required
for male sex development
Vitamin
D is reouired
for bone
develooment
Retinoic
acid
is
a
morphogen
Thyroid
hormones
Thyroid hormones
control basal metabolic
rate
II
,oz\o4.",-8:o'
\_/
v
'ilH
I
triiodothyronine
(T.)
tl
u^ | |
i?"^7---,y'cu"
F'l
eU I
-HC
tl
CH
c-cH"
il-
HC
CH
tl
HC
c-cH.
il"
HC
cooH
(trans)
retinoic
acid
!:i,i,q:i:1
.11 l::.
SeveraI
types of hydrophobic
smat[
motecutes
activate tran-
scription factors.
The
adrenal
gland
secretes
>30
steroids,
the
two major
groups
being the
glucocorticoids
and
mineralocorticoids.
Steroids
provide
the repro-
ductive
hormones
(androgen
male
sex
hor-
mones and
estrogen female
sex
hormones).
Vitamin
D is required for
bone development.
Other
hormones, which
have unrelated
structures
and
physiological purposes,
function
at the
molecular level in
a similar
way to the
steroid
hormones. Thyroid
hormones,
which
are
based on iodinated
forms of
tyrosine, con-
trol basal metabolic
rate in
animals. Steroid
and
thyroid
hormones
also may be important
in
metamorphosis (ecdysteroids
in insects
and thy-
roid hormones
in frogs).
Retinoic acid
(vitamin
A) is
a morphogen
responsible for development of the anterior-pos-
terior axis in the developing chick limb
bud.
Its
metabolite, 9-cis
retinoic
acid, is found in tis-
sues that are major sites for storage and metab-
olism of vitamin A.
We may account
for
these various actions
in terms
of
pathways
for
regulating
gene
expression. These
diverse compounds share a common mode
of
action: Each is a small molecule that binds t0 a
spe-
cific
receptor
that
activates
gene
transcription.
("Receptor"
may be a
misnomer:
The
protein
is a receptor for steroid or thyroid hormone in
the same sense that /ac repressor is
a
receptor
for
a
p
galactoside,
i.e., it is not
a receptor in the
sense of comprising a membrane-bound
pro-
tein that
is
exposed to the cell surface.)
Receptors for the diverse
groups
of steroid
hormones, thyroid hormones, and retinoic
acid
represent
a
new
"superfamily"
of
gene
regu.ta-
tors, the ligand-responsive activators. AII
the
receptors have independent
domains for DNA-
binding and hormone binding that
are
in
the
same relative locations. Their
general
organi-
zation is
summarized
in f:{j*fti
tli,: {i.
The central
part
of the
protein
is
the DNA-
binding
domain.
These regions
are closely
related
for the
various
steroid receptors
(from
the most closely related
pair,
wilh94oh sequence
identity, to the least well related
pair,
at 42"/o
identity). The act
of binding
DNA
cannot be dis-
connected from
the ability to activate transcrip-
tion, because mutations in
this domain affect
both activities.
The
N-terminal
regions
of the receptors
show
the least
conservation of sequence. They
include
other
regions
that are needed to
activate
transcription.
The C-terminal
domains bind the hor-
mones.
Those in the steroid receptor
family
show identities ranging from
30oh ro 57oh,
reflecting
specificity for individual
hormones.
Their
relationships with the
other receptors
are minimal
and
reflect
specificity for
a vari-
ety of compounds-thyroid
hormones.
vita-
min D, retinoic
acid, and so forth.
This domain
also has the motifs responsible
for dimeriza-
tion and a region involved
in transcriptional
activation.
Some ligands
have multiple receptors
that
are closely related,
such as the three
retinoic
acid receptors
(RARo,
p,
and
y)
and the
three
receptors for 9-cis-retinoic
acid
(RXRa,
B,
and
y).
CHAPTER 25 Activating
Transcription
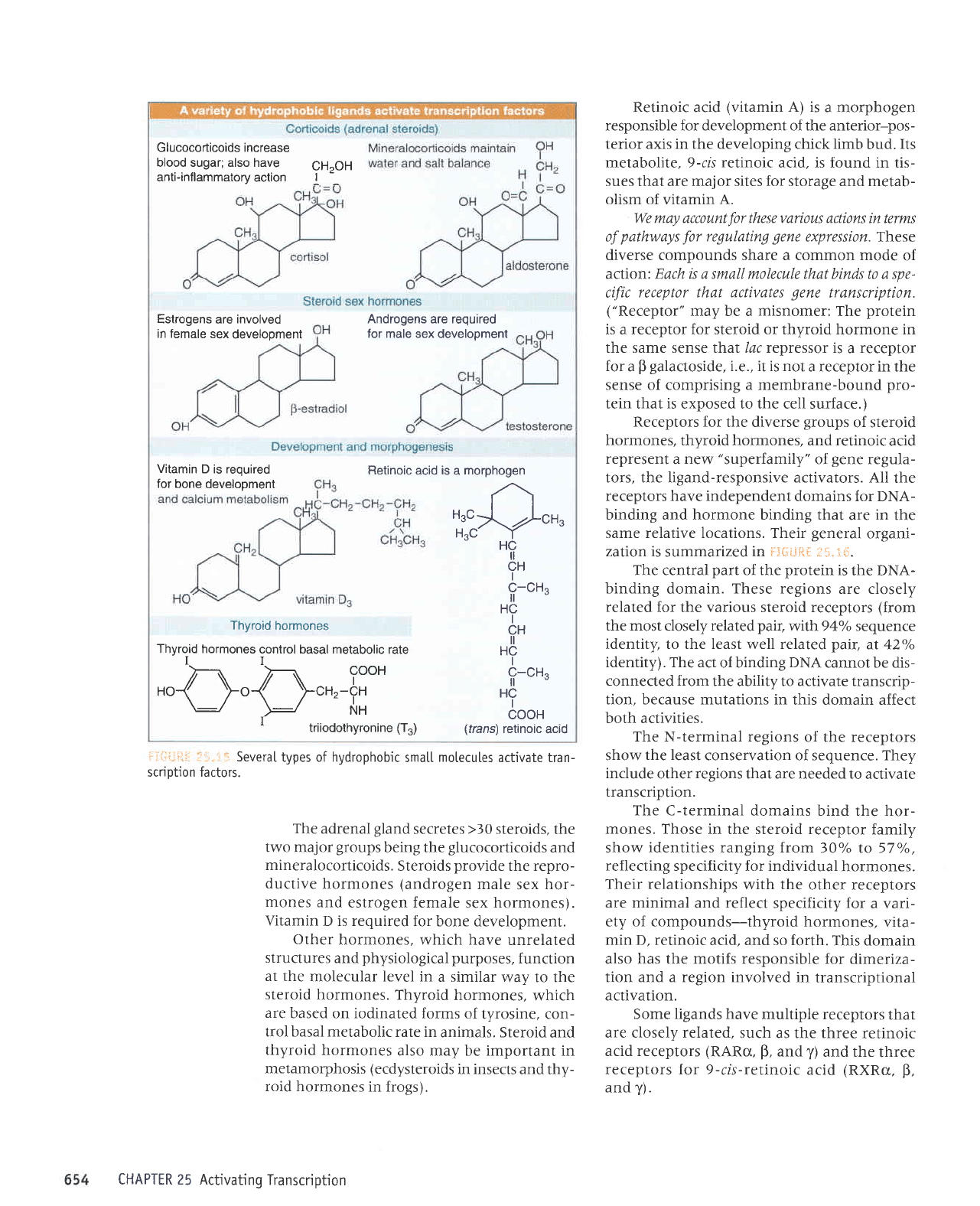
transcription)
Hormone-binding
regions
and
dimerization
(identity
varies from
57or"-15q")
Glucocorticoid
57 Mineralocorticoid
65 Pr^daaiar^na
Androgen
Estrogen
Triiodothyronine
Vitamin
D
Retinoic
acid
9-cls Retinoic acid
l:.ir::,i+ii ,r-1, i;l Receptors for
many
steroid and thyroid hor-
mones have a similar
organization, with
an
jndividuaI
N-termjnaI region,
conserved DNA-binding region,
and a
C-terminal hormone-binding region. Identities
are relative
to GR.
Steroid Receptors
Have Tinc
Fingers
.
The DNA
binding domain of a steroid receptor
is a
type of
zinc finger
that has
Cys but not His
residues.
.
Glucocorticoid and
estrogen receptors
each
have
two zinc fingers, the first
of which determines
the
DNA target sequence.
r
Steroid receptors
bind to DNA as dimers.
Steroid receptors
(and
some
other
proteins)
have another
type of zinc finger
that
is
differ-
ent
from
Cys2/His2 fingers. The
structure is based
on a sequence with the zinc-binding
consensus:
Cys-X2-Cys-X1
3-Cys-X2-
Cys
These
sequences are
called Cys2/Cys2 fin-
gers.
Proteins with Cys2/Cys2 fingers
often have
nonrepetitive fingers, in
contrast with the tan-
dem repetition
of the Cys2/Hisz type. Binding
sites in DNA
(where
known)
are short and
palindromic.
The
glucocorticoid
and estrogen receptors
each
have two fingers,
each with a zinc atom at
the center of a tetrahedron
of cysteines. The
two fingers form o,-helices
that fold together to
:S
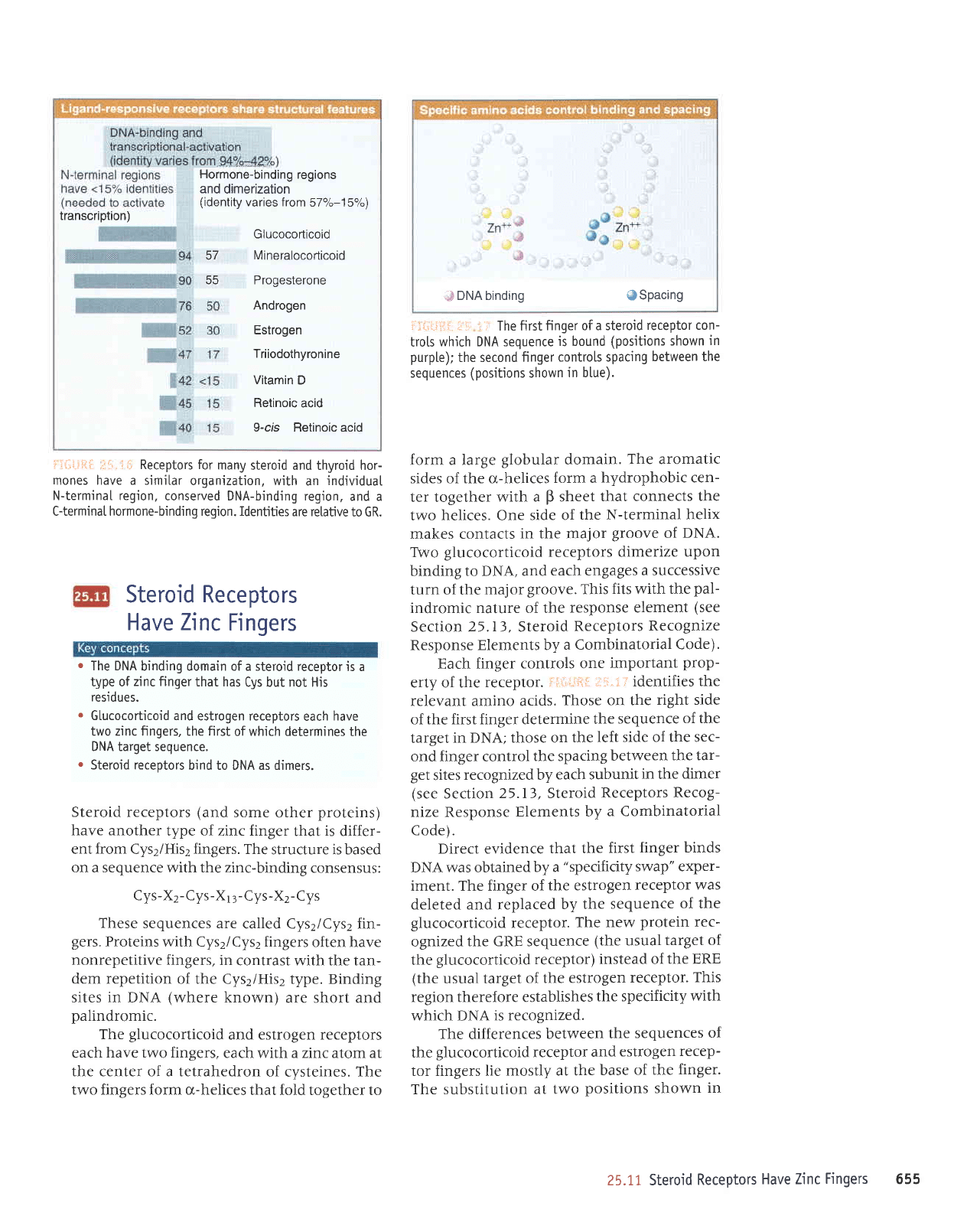
DNA binding
@Spacing
i',ir.:i-jiii
:jl l
: The firstfinger
of a steroid
receptor con-
trots
whjch DNA sequence
is bound
(positions shown
in
purpl.e);
the second finger
contro[s spacing
between the
sequences
(positions
shown
in b[ue).
form a large
globular
domain.
The aromatic
sides of the cr-helices
form a
hydrophobic cen-
ter together
with a
p
sheet
that connects
the
two
helices.
One
side of
the N-terminal
helix
makes contacts
in the major
groove
of
DNA.
TVvo
glucocorticoid receptors dimerize
upon
binding to DNA, and
each engages
a successive
turn of the
major
groove.
This
fits
with
the
pal-
indromic nature of the
response element
(see
Section 25.I3, Steroid
Receptors
Recognize
Response Elements by
a Combinatorial
Code).
Each finger
controls one
important
prop-
erty of the
receptor.
i'ir.iitfiir
iiir.i i' identifies
the
relevant amino acids.
Those on the
right side
of the first finger determine
the
sequence
of the
target
in DNA; those on
the
left side of the sec-
ond finger control
the spacing
between
the tar-
get
sites
recognized by
each subunit
in
the
dimer
(see
Section
25.1), Steroid
Receptors
Recog-
nize Response
Elements by
a Combinatorial
Code).
Direct evidence
that the
first
finger binds
DNA was obtained
by a
"specificity
swap" exper-
iment. The finger of
the estrogen
receptor
was
deleted and replaced
by the
sequence
of the
glucocorticoid
receptor.
The
new
protein
rec-
ognized
the GRE sequence
(the
usual
target of
the
glucocorticoid receptor)
instead of
the ERE
(the
usual target
of the
estrogen
receptor.
This
region therefore establishes
the specificity
with
which DNA
is recognized.
The differences
between
the
sequences
of
the
glucocorticoid
receptor
and
estrogen
recep-
tor fingers lie
mostly at the
base
of the
finger.
The substitution
at two
positions
shown
in
25.11 Steroid
Receptors
Have Zinc
Fingers 655

i:al-=!
ii.t* Discrimination
between
GRE and
ERE
tar-
get
sequences is
determined by two amino
acjds at the
base of the first zinc finger
in the receptor.
Same sequence
in
both
receptors
Different
sequence
in
each
receptor
ttG
Ln
^^s
GRE
ERE
specif
icity
sPecif icitY
GRE/Enhancer
Promoter
We know the most about the interaction
of
glu-
cocorticoids with their receptor, whose
action
is illustrated in Fii:ii.:trF. itir-3*.
A steroid hormone
can
pass
through the cell membrane
to enter
the cell by simple diffusion. Within
the cell, a
glucocorticoid
binds the
glucocorticoid
receptor.
(Work
on the
glucocorticoid
receptor
has relied
on the synthetic steroid
hormone,
dexametha-
sone.) The Iocalization of free receptors
is not
entirely clear; they may be in equilibrium
between the nucleus and cytoplasm.
When hor-
mone binds to the receptor.
though, the
pro-
tein is
converted
into
an activated form that has
an increased affinity for DNA, so the hormone-
receptor
complex
is
always
localized
in the
nucleus.
The
activated receptor recognizes a
specific
consensus sequence that identifies
the GRE.
The GRE is typically located in
an enhancer that
may
be several
kb
upstream or downstream
of
the
promoter.
When the steroid-receptor
com-
plex
binds to the enhancer, the nearby
promoter
is activated and transcription initiates
there.
Enhancer activation
provides
the
general
mech-
anism by which
steroids
regulate
a wide
set of
target
genes.
The C-terminal region regulates
the activ-
ity of the receptor in a way that
varies for the
individual receptor. If
the C-terminal domain
of the
glucocorticoid
receptor
is deleted,
the
remaining
N-terminal
protein
is constitutively
active:
It
no longer requires steroids for
activ-
ity. This suggests that, in the
absence
of steroid.
the steroid-binding domain
prevents
the recep-
tor from recognizing the GRE; it
functions as
an internal negative regulator. The
addition of
steroid inactivates
the
inhibition,
releasing
the
receptor's ability
to bind the GRE and
activate
transcription. The basis for the repression
could
be
internal,
relying on interactions
with another
part
of the receptor,
or
it
could result from
an
interaction with
some other
protein
that is
dis-
placed
when steroid binds.
The interaction between
the domains is
dif-
ferent in
the estrogen receptor. If
the hormone-
binding domain is
deleted, the
protein
is unable
to activate
transcription, although it
continues
to bind to the ERE. This
region is
therefore
required
to activate rather
than to repress
activity.
+:Ii;.= +:;:
:l
i.
.t
*
G lucocorticoids regutate
gene
transcrip-
tion
by causing their receptor
to bind
to an enhancer
whose
action is needed for
oromoter
function.
ir:,i.iitI
lil..rt:
allows
the
glucocorticoid
receptor
to bind at
an ERE instead
of a GRE.
@
Binding
to the Response
Element
Is Activated
by
Ligand-Binding
.
Binding
of Ligand
to the C-terminaI
domain
increases
the affinity
of the DNA-binding
domain
for its
specific target site in DNA.
656
CHAPTER
25 Activating
Transcription
Steroid
Receptors
Recognize
Response
Elements
by a
CombinatoriaI
Code
.
A
steroid
response
element consists
of two short
half sites that may
be
palindromic
or directly
repeated.
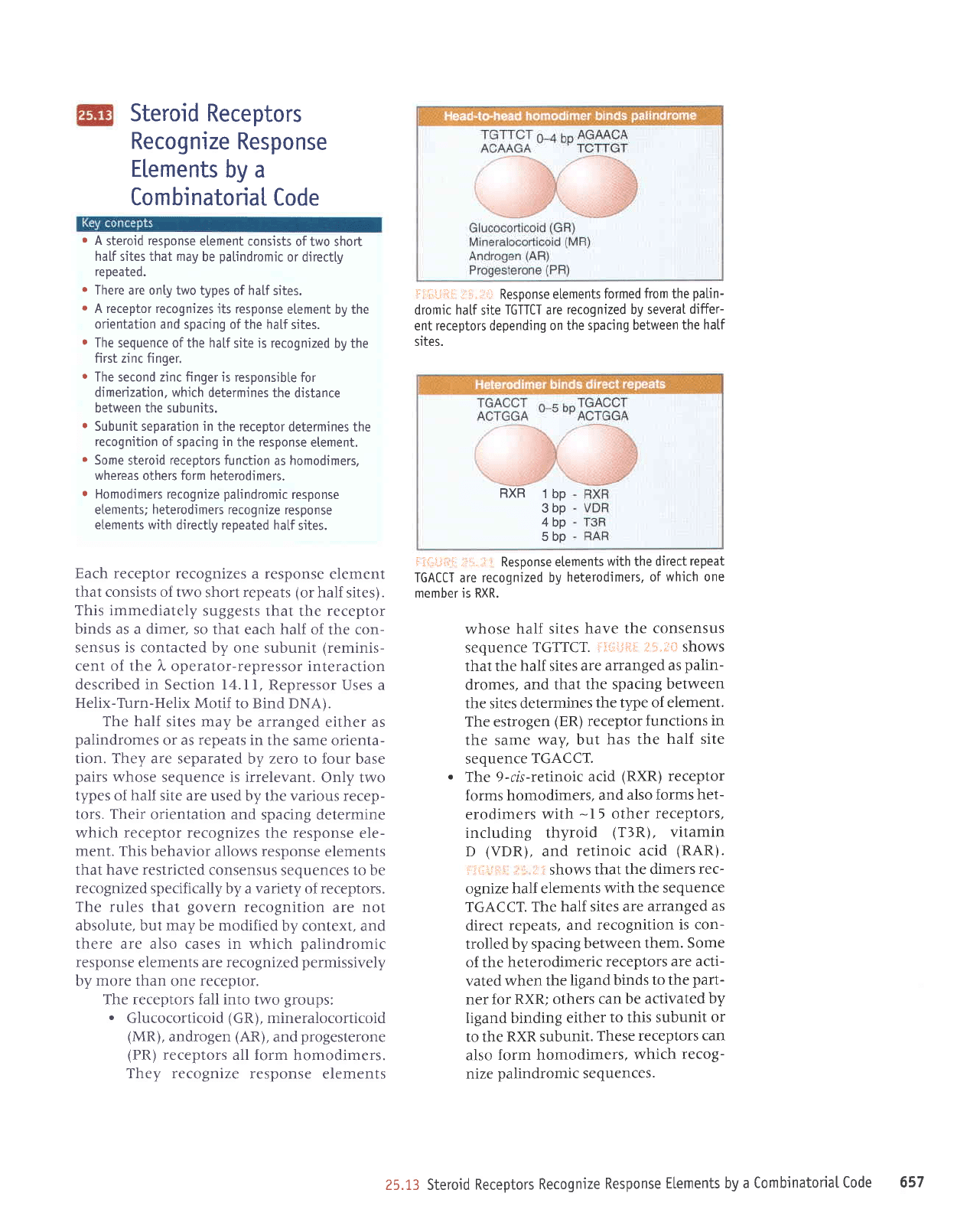
r
There are onty two
types of half sites.
r
A receptor recognizes
its response
etement by the
orientation
and spacing of
the
ha[f
sites.
.
The sequence
of the
half
site is recognized
by the
first zinc finger.
r
The second zinc finger is
responsible for
dimerization, which
determines the
distance
between the subunits.
r
Subunit separation in
the receptor
determines the
recognition of spacing in
the response
etement.
r
Some
steroid
receptors
function
as
homodimers.
whereas
others
form
heterodimers.
r
Homodimers
recognize
pa[indromic
response
etements;
heterodimers
recognize response
etements with
directly repeated hatf
sjtes.
Each receptor recognizes
a response
element
that consists of two
short repeats
(or
half
sites).
This immediately
suggests
that the receptor
binds
as a dimer, so that
each half of the con-
sensus is contacted
by one subunit
(reminis-
cent of the l"
operator-repressor interaction
described in Section l4.ll,
Repressor Uses a
Helix-Ti-rrn-Helix
Motif to Bind DNA).
The half
sites may be arranged
either as
palindromes
or as repeats in
the same orienta-
tion. They are separated
by zero to four base
pairs
whose
sequence is irrelevant.
Only two
types of half site are
used by the
various
recep-
tors.
Their
orientation and
spacing determine
which receptor recognizes
the response ele-
ment. This behavior
allows response elements
that
have restricted
consensus
sequences to be
recognized specifically
by a variety of receptors.
The rules
that
govern
recognition
are
not
absolute, but may be modified
by context, and
there are also cases in which
palindromic
response elements are recognized
permissively
by
more than
one
receptor.
The receptors fall into
two
groups:
.
Glucocorticoid
(GR),
mineralocorticoid
(MR),
androgen
(AR),
and
progesterone
(PR)
receptors all form
homodimers.
They recognize
response elements
lii:ijiii-:
i:.'r,,l,ri:
ftg5p6n5e
etements
formed
from
the
patin-
dromic hatf
site
TGTTCT are
recognized by several
differ-
ent
receptors
depending
on the spacing
between the
hatf
sites.
ii{,;Lii,;l: ,iii..,:
i Response
etements
with the direct
repeat
TGACCT are recognized
by
heterodimers, of
which one
member is RXR.
whose
half sites
have the
consensus
sequence
TGTTCT.
fl{iiiiiiJ
;:*,ili-l shows
that the half
sites are
arranged as
palin-
dromes,
and that
the spacing
between
the sites determines
the tlpe of element.
The estrogen
(ER)
receptor functions
in
the same
way, but
has the
half site
sequence
TGACCT.
.
The
9-crs-retinoic
acid
(RXR)
receptor
forms homodimers,
and
also forms
het-
erodimers
with
-15
other
receptors,
including thyroid
(TlR),
vitamin
D
(VDR),
and
retinoic
acid
(RAR).
i llt,
1.r
r.;
I ;,f
..i,i
shows
that the dimers
rec-
ognize
half elements
with
the sequence
TGACCT.
The
half sites are
arranged
as
direct repeats,
and
recognition
is con-
trolled by
spacing
between them.
Some
of the heterodimeric
receptors
are acti-
vated
when the
ligandbinds
to the
part-
ner for RXR;
others
can be activated
by
ligand binding
either
to this subunit
or
to the
RXR subunit.
These
receptors
can
also
form homodimers.
which
recog-
nize
palindromic sequences.
RXR
1
bp
3bp
4bp
5bp
25.13 Steroid Receptors
Recognize
Response
Elements by
a Combinatorial
Code 657

Now we
are
in
a
position
to understand
the basis for
specificity of recognition. Recall
that Figure 25.17
shows how recognition of
the
sequence of the half site is conferred
by
the amino acid sequence in
the first
finger.
Specificity for
the spacing between half sites
is carried
by amino acids in the second finger.
The
structure of the dimer determines
the dis-
tance between the
subunits that sit in succes-
sive turns
of the major
groove,
and thus
controls
the response to
the spacing of half
sites. The exact
positions
of the residues respon-
sible for
dimerization differ in individual
pair-
wise combinations.
How
do the steroid receptors
activate tran-
scription? They
do not act directly
on the basal
apparatus,
but rather function
via a coactivat-
ing
complex. The coactivator
includes various
activities,
including the common
component
CBP/p300, one of whose functions
is to mod-
ify
the structure
of chromatin
by acetylating
histones
(see
Figure 30.I4).
AII receptors
in the superfamily
are
ligand-
dependent
activators
of transcription, but some
are also able to repress
transcription. The TR
and RAR receptors,
in the form
of
heterodimers
with RXR,
bind to certain loci
in the absence of
ligand
and repress
transcription by means
of
their ability
to interact with a
corepressor
pro-
tein. The
corepressor functions
by the reverse
of the mechanism
used by
coactivators: It
inhibits
the function of the
basal transcriotion
--:i,UliI
i:'!.1]!' The
steroid receptors
TR and RAR
bind the
SMRT corepressor
in the
absence of ligand. The
promoter
is not
expressed. When
SMRT is disptaced
by binding of ti-
gand.
the receptor
binds a coactivator
comptex. This
leads
to activation
of transcription
by the basaI
apparatus.
CHAPTER
25 Activating
Transcription
apparatus, one of its actions being the deacety-
Iation of histones
(see
Figure 30.16). We do not
know the relative importance of the repressor
activity vis-d-vis the ligand-dependent
activa-
tion
in
the
physiological
response
to hormone.
The effect of ligand binding
on the re-
ceptor is to convert it from a repressing
com-
plex
to an activating complex, as shown in
FlSlJft[ :S.I*.
In the absence of ligand, the recep-
tor is bound to
a corepressor complex. The com-
ponent
of the corepressor that binds
to the
receptor is
SMRT.
Binding
of
ligand
causes a
conformational change that displaces SMRT.
This
allows the coactivator to bind.
Homeodomains Bind
Related
Targets
in DNA
.
The homeodomain is
a
DNA-binding
domain
of 60
amino
acids that
has
three cr-helices.
r
The C-terminaI
q-helix-3
is 17
amino acids
and
binds in
the
major
groove
of DNA.
o
The N-terminal
arm of the homeodomain
projects
into the minor
groove
of
DNA.
o
Proteins
containing homeodomains may
be either
activators or repressors of transcription.
The homeobox is
a sequence that
codes
for
a
domain
of 60 amino acids
present
in
proteins
of
many
or even all eukaryotes. Its name
derives
from its original identification
in Drosophila
homeotic loci
(whose genes
determine
the iden-
tity of body
structures).
It is
present
in many
of
the
genes
that
regulate
early development
in
Drosophila,
and a related motif is
found in
genes
in
a wide range of higher eukaryotes.
The ho-
meodomain
is found in many
genes
concerned
with
developmental regulation.
Sequences
related
to the homeodomain
are found in
sev-
eral types of animal
transcription factors.
In
Drosophila homeotic
genes,
the homeo-
domain often
(but
not
always) occurs
close to
the
C-terminal end. Some
examples
of
genes
containing homeoboxes
are
summarized
in
F:fu{jqg ;}.5.f 3.
Often the
genes
have little
con-
servation of sequence
except in the homeobox.
The
conservation
of
the
homeobox
sequence
varies. A major
group
of homeobox-contain-
ing
genes
in Drosophila
h'as a
well conserved
sequence,
with
80% Io 90o/o similarity
in
pair-
wise
comparisons. Other
genes
have less
closely
related
homeoboxes. The
homeodomain
is
sometimes
combined with
other motifs in
ani-
mal
transcription factors.
One
example is
pre-
Corepressor
Steroid receotor
Ligand
sented by the
Oct
(octamer-binding)
proteins,
in
which a conserved
stretch
of
75
amino acids
called the Pou region
is located
close
to a region
resembling
the homeodomain.
The
homeoboxes
of the Pou
group
of
proteins
are the least
closely
related
to the original
group,
and thus comprise
the farthest extension
of the
family.
The homeodomain
is responsible
for
bind-
ing
to DNA, and
experiments
to swap ho-
meodomains
between
proteins
suggest
that the
specificity of DNA recognition
lies within
the
homeodomain.
As
with
phage
repressors,
though, no simple
code relating
protein
and
DNA sequences
can
be deduced. The
C-terminal region of the homeodomain
shows
homology
with the helix-turn-helix
motif of
prokaryotic
repressors.
We recall from
Sec-
tion 14. I l, Repressor
Uses a Helix-Turn-Helix
Motif to Bind DNA,
that the l,
repressor has
a
"recognition
helix"
(a-helix-3)
that makes
con-
tacts in the major
groove
of DNA,
whereas the
other helix
(u-helix-2)
lies at
an angle across
the
DNA.
The homeodomain
can
be organized
into three
potential
helical
regions;
the
sequences of three
examples are
compared in
f;.ii-in-il
f :1.i'... The
best conserved
part
of the
sequence lies in
the third helix.
The difference
between these structures
and the
prokaryotic
repressor
structures lies in
the length
of the
helix that recognizes
DNA, helix-3,
which is l7
amino acids long in
the homeodomain,
com-
pared
to nine residues
long in
the
l,
repressor.
The
structure of the homeodomain
of the
D. melanogasler
engrailed
protein
is represented
schematically in
Fll"irJlti:
;15"i":fl. Helix 3 binds
in
the major
groove
of DNA and
makes the major-
ity
of the contacts between
protein
and
nucleic
acid. Many of the contacts
that orient the
helix
in the
major
groove
are made with
the
phos-
phate
backbone, so they are
not specific for DNA
sequence. They lie largely on
one face of the
double helix,
and
flank the bases
with which
specific contacts are
made. The remaining
con-
tacts
are made by
the N-terminal
arm of the
homeodomain, the sequence
that
just
precedes
t:T{-,iiltt
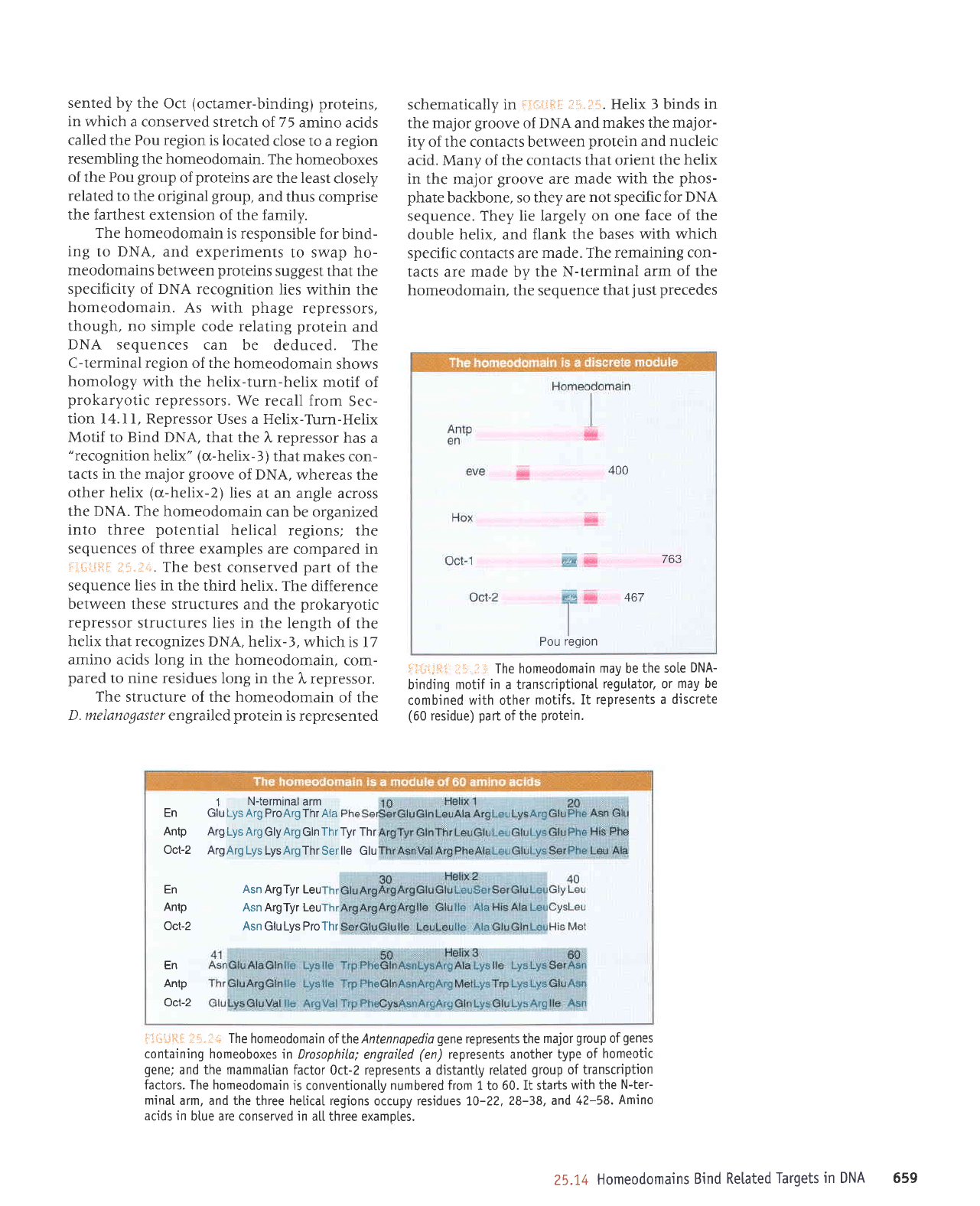
;ilii.il:i The homeodomain
may
be
the so[e
DNA-
binding motif in a transcriptionaI
regulator. or
may be
combined with other
motifs. It
represents a discrete
(60
residue)
part
of the
protein.
Flii'js.[
]i.;i;;
The
homeodomain
of the
Antennapedio
gene
represents the
major
group
of
genes
containing homeoboxes
in Drosophila;
engroiled
(en)
represents another type
of
homeotic
gene;
and the mammatian
factor 0ct-2 represents a distantly retated
group
of transcription
factors. The homeodomain
is
conventionatly
numbered from 1
to
60. It starts
with the
N-ter-
minaI
arm, and the
three helicaI
regions
occupy residues 70-22,
28-38, and 42-58.
Amino
acids in
btue are conserved in
atl three examotes.
400
763
-'i:'
467
I
I
I
Pou
region
En
Antp
Oct-2
En
Antp
Oct-2
En
Antp
Oct-2
1
N-terminal arm
Glu
Lys Arg ProArgThr Aia
Arg Lys Arg
GlyArg Gln
Thr Tyr
Thr
ArgArg Lys
LysArgThr Serlle Glu
Asn
ArgTyr Leu
Asn ArgTyr Leu
Asn GluLys ProThr
25.14
Homeodomains
Bind Retated
Targets
in DNA 659

N-terminal
arm
lies
Helices
1 and 2 lie above the DNA
In mrnor
groove
Helix
3
lies in
the major
groove
.rt::
i:L
:
.
::
Hetix 3 of the homeodomain
binds
in
the
major
groove
of DNA, wjth helices 1 and 2
lying outside the
double hetjx. Hetix
3 contacts both the
phosphate
backbone
and specific bases. The N-terminaI
arm hes in the minor
groove,
and makes
additionaI contacts.
the first
helix. It
projects
into
the
minor
groove.
Thus
the N-terminal and
C-terminal regions of
the homeodomain
are
primarily
responsible for
contacting
DNA.
A striking
demonstration of the
generality
of this model
derives from a comparison
of the
crystal
structure of the homeodomain
of
engrailed protein
with
that o{ the o2 mating
protein
of
yeast.
The
DNA-binding domain
of
this
protein
resembles a homeodomain
and can
form
three similar helices:
Its structure in the
DNA groove
can be superimposed almost
exactly
on that of the
engrailed homeodomain. These
similarities
suggest that all homeodomains
bind
to DNA in
the same manner. This
means that a
relatively
small
number
of residues in helix-3
and in the
N-terminal arm are responsible
for
specificity
of contacts with DNA.
One
group
of
homeodomain-containing
proteins
is
the set of Hox
proteins.
They bind
to DNA
with rather
low sequence
specificity,
and it has
been
puzzling
how
these
proteins
can
have
different specificities.
It turns
out
that Hox
proteins
often bind
to DNA as heterodimers
with a
partner (called
Exd
in flies and Pbx in
vertebrates).
The heterodimer
has
a
more
restricted
specificity in vitro
than an individual
Hox
protein;
typically
it binds
the l0 bp
sequence
TGATNNATNN.
This
still
is
not enough
to account
for the differences
in the
specificities
of Hox
proteins.
A third
protein,
Hth,
which is
necessary
to localize Exd
in the nucleus,
also
forms part
of the complex
that binds DNA,
and
may restrict
the
binding sites further.
The same
partners
(Exd
and Hth)
are
present
together
CHAPTER 25
Activating
Transcription
with each
Hox
protein
in
the trimeric complex,
though, so it remains
puzzling
how each Hox
protein
has
sufficient
specificity.
Homeodomain
proteins
can be either tran-
scriptional activators or
repressors.
The nature
of
the factor depends on the other domain(s)-
the homeodomain
is responsible
solely for bind-
ing to DNA. The activator or repressor
domains
both act by influencing the basal apparatus.
Activator domains may
interact
with coactiva-
tors that in turn bind to components of the
basal
apparatus.
Repressor
domains also
interact
with
the transcription apparatus
(that
is,
they do not
act by blocking access to
DNA
as such). The
repressor Eve, for example, interacts directly
with TF'D.
He[ix- Loop-
Helix
Protei ns
Interact
by CombinatoriaL
Association
.
Hetix-toop-helix
proteins
have
a motif of 40
to 50
amino acids that comprises two amphipathic
c
helices of 15 to 16 residues separated
by a [oop.
.
The hetices
are
responsibte for
dimer
formation.
.
bHLH
proteins
have
a basic sequence adjacent to
the HLH motif
that
is responsible for
binding to
DNA.
.
Class
A
bHLH
proteins
are ubiquitousty
expressed.
Ctass B bHLH
proteins
are tissue-specific.
r
A class B
protein
usualty forms
a
heterodimer
with
a ctass A
protein.
r
HLH
proteins
that lack the basic region
prevent
a
bHLH
partner
in a heterodimer from
binding to
DNA.
r
HLH
proteins
form
combinatorial associations
that
may
be changed during development
by the
addition
or
removaI
of specific
proteins.
TWo common features in
DNA-binding
pro-
teins are the
presence
of
helical
regions
that
bind
DNA
and the ability of the
protein
to
dimerize. Both features are represented
in the
group
of helix-loop-helix
(HLH) proteins
that
share a common type
of sequence motif: A
stretch of 40 to 50 amino
acids contains
two
amphipathic o helices separated
by a linker
region
(the
loop)
of varying length.
(An
amphi-
pathic
helix
forms two faces,
one
presenting
hydrophobic
amino acids, and the
other
pre-
senting
charged amino acids.) The
proteins
in
this
group
form
both homodimers
and het-
erodimers
by
means
of interactions
between
the hydrophobic
residues
on the correspon-
ding faces
of the two helices. The helical
regions
