Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.

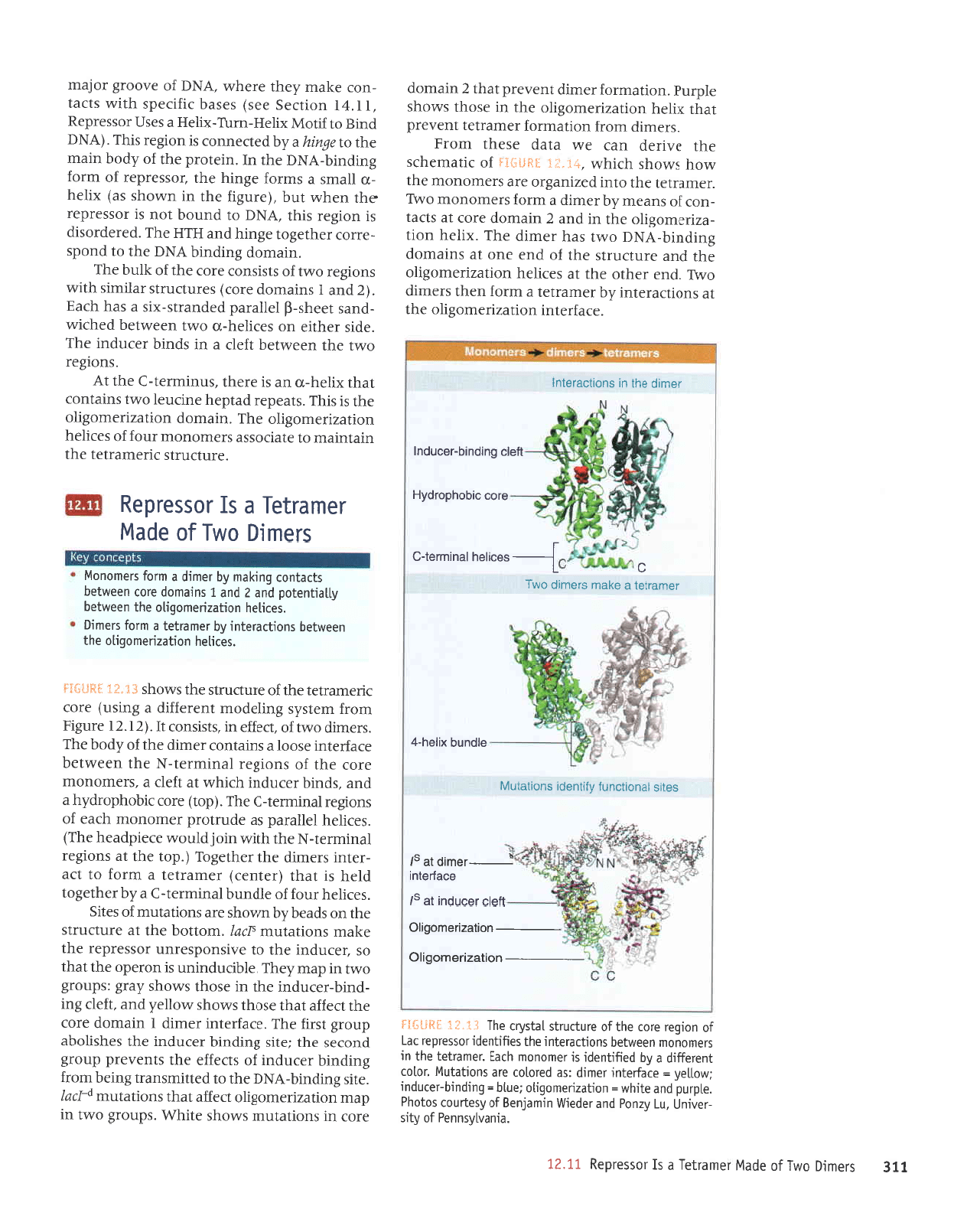
II€
sraur0
oMl
Jo
apPW
lauellaf
P sI lossaldau
II'zI
'Prue^l^suuad
Jo
^lrs
-ralrun
'nl
r\zuo4 pue
tapatl4
ururefueg
1o
r\salnor
so1oq6
'aldrnd
pue
eJt[lm
=
uor]pzuauo6Uo
ianlq
=
6urpurq-lalnpur
1rvro11aA
=
alpJlalur
.laurp :sp
pe.lolol
olp
suoqplnl^I
.lolol
luoia#rp
e
fq parlrluapr
sr loutouout qlpl
:laupllal
aql
ur
sraurououl
uoamlaq
su0qlplelur
aql soJJquapr
lossardal
re1
1o
uor6ar
alol aql
Jo
alnllnlls
1ep&r
eq1
f
i..f;1
3g{l:ilj
uotlezueulobr;g
nezueuoOrlg
ualc
lecnpu!
lE
s/
ocPlolul
-laultp
le
s/
salrs
leuotpunl
A;uuapl
suorlelnyl
olpunq xrlor.l-t
reuJellel
e olPu.r
sJorutp
oMI
|."r',"r.{
reururot-c
eroc
crqoqdotplg
yelc
6urpurq-recnpu;
raurp
aql
ul suotlc€Jolul
'Jf,eJJelur
uorlezrJaruoSlo
aql
le
suorlJeralur
dq
raurerlel
p
uroJ
uaql
sraurp
o^41
'puJ
JJqlo
aql
le
sJlrlaq
uorlezrJJruoSrlo
JI{l
pue
JJnl)nrls
Jql
Jo
puJ
Juo
lp
sureurop
Surpurq-y1qq
oMt seq
rJrurp
JqJ
.xrlJq
uorl
-ezrJeruoSrlo
Jql
ul
pue
Z
ureuop
aJoJ
lp
sDel
-uoJ
Jo
supaur
Lq rarurp
p
ruJoJ
sJJurouour
ol4l
'JJLUeJIJI
Jql
01ur
pazrueSro
JJe sJauouotu
aql
Moq
sMoqs qJIqM
'!i.";j{
:sf]*l+
Jo
JrlpruJqJs
aql
a^rrap
uPf,
Jl!|
elpp
Jsaql
ruoJd
'sJJurrp
uroJJ
uorlpurJoJ
rerueJlel
luaaard
fpqt
xleq
uollezlJJruoS[o
aql
ur Jsoql
s,,lroqs
a1drn4
'uorleurroJ
Jarurp
luJAaJ
dWr+
T.
urpruop
aJoJ ur suorlplnu
smoqs Jllq6'sdnor8
o,lrl ur
deur
uorlezrrauro8r1o
TJJJJe
lpqr
suorlelntu
p_1rrl
'a1rs
Surpurq-VNq
Jqt
ot
pJltrrusuprt
Suraq ruor;
Surpurq
JaJnpur
Jo
sDalJe
aq1
sluaaard
dnor8
puoJes
Jql
:Jlrs
Supurq rJJnpu
aq1 saqsrloqe
dnor3
lsrg
aqJ
'e)eJretur
Jarurp
I
uretuop
eJo)
rql
irrJ;e
teql
rsoqt
sMoqs
uo11a,{.
pue
'1;ap
8ur
-purq-JeJnpu
eq1 ur
esoql s.&tor{s
LerS :sdnorS
oml ur deru
daql
'elqDnpurun
sr uoJedo
Jql
leql
os teJnpur
aql
ol e^rsuodseJun
JossJJdeJ
eql
a>leru
suorletnut
{)ol'Iuolloq
aql
]e
erntJnJls
Jql
uo speaq lq
uatoqs
aJe suouelnru
Jo
salrs
'saJrIJq
JnoJ
Jo
elpunq
lpururJl-)
e.,(q
raqlaS0l
plaq
sI
teql
(JatuaJ)
raruerlal
p
uroJ
ot
1re
-Jalur
srarurp
Jql
raq1e3o1
('dol
aql
te
suo€ar
IpurruJal-N
Jql
r{lrm urotplno,ra
a;ardpeaq
aql)
'se)Ileq
lalpred
se
apnrl0rd
Jeruouour q)pJ
Jo
suor8ar
punuret-l
aqI'(dor)
aror
trqoqdorpz(q
e
puP
'spurq
JaJnpur
q)lq^{
]e
uJIl
p
'sJerrrouoru
JJoJ
eql
yo
suor8ar
leurrrrJJt-N
eql
uJaMleq
aJpJJalu
Jsool
p
surpluoJ
JJrurp
aqt
yo,{poq
aql
'sJalrrp
o.ttt
Jo
'DeJJa
ur
'slslsuoJ
lI'(ZI.U
arn8rg
ruor;
ura1s.{s
Surlaporu
tuJJeJJrp
e
Sursn)
aror
JUJIIIPTIJ]
Jql
Jo
arnDru$
aql
sMoqs
f
g.'a'i
Isfl*3j
'satllaq
uotlezuauoEqo
aq1
uaaMlaq
suorllplalur
fq
tauerlal
p
uloJ slaurr0
'saluoq
uoqpzuauo6rlo
aLll uao/v\laq
fllequalod pup
Z
pup
I
sureuop
arol ueamlaq
sllpluol
6uqeu
[q laulp
e
ulloJ siauouo14
srauro
oMI
Jo
apP|/{
lauPllal
P
sJ tossaldau
'eJnlJnJls
JrJJrueJlal
aql
urPlureru
01 Jlpl)osse
sJaruouoru
rnoJ
Jo
seJrleq
uorlezrJJruoSrl0
aq1'urerxop
uorlezrraruoSrl0
Jql sI
sIqJ'steadeJ ppldeq
JurJnal
o,vrt
sulpluoJ
leq]
xrlaq-?o
up sr
JJar{l
'snurruJal-)
Jql
ly
^
'SUOItsEJ
o1!tl
aql
uaJMlJq
lJelJ
e ur spurq
JaJnpur
aqJ
'Jprs
rJr{lrJ
uo
seJrlaq-n
ol!1l
uJaMlJq paq)r^t
-pues
laaqs-(
lalpred
prpuels-xrs
p
spq
qlpf
'(Z
pup
I
surpuop
aror) sarnpnJls
lelrurs qlrm
suor8ar
omt
Jo
stsrsuoJ
JroJ
ar{t
Jo
{lnq
JqJ
'ureruop
Surpurq
yN(
aq1
o1
puods
-eJrof,
raq1a3o1
aSurq pue
HJH
eqJ
.perepJosrp
sr
uor8ar
s1ql
'VNCI
01
punoq
tou
sr rossardar
rqt
ueqM
1nq
'(arn8r;
eqt
ul uMoqs
se) x11aq
-xl
flprus
e sruJoJ
a8urq
aqt
?ossardar
Jo
ruroJ
Surpurq-ypq
Jql
uI
'uralord
aql;o,{.poq
ureur
aqt
o1 a6uu7
e dq
palrauuol
sr uor8ar
slqJ
'(VNq
pulg
ot
JIIOW
xrlaH-urnJ-xrlaH
p
sasn
Jossarda6
'IlvI
uorlJJS
aas) saseq
tr;roads qllM
sltel
-uoJ
e>lpu z(aqt
araq,u
'VN61
Jo
anoor8
roleru
'lrunqns
lossoroar aql
Jo
sutPtuop
lualo#rp
ur
rnrro
suoqelnu
Jo
sad^l
]uoraJJr[
o
aJnllnjls ur.Pulo0
eLJl
q]!M
elPleilol
sadAlouaqd
tuelny11
@
'roterado
Jqt roJ .dlrur11e eqt seJnpal
pue
:ossardJr JrJarurllnu aqt
yo
a8eluenpe
aqt
sJleurullJ slqJ'dlsnoauetlnuls
VNC
pulq
ra8uol
ou ueJ JJrurp e ur sarardpeaq o,t,rl eql
]Pq1 llnseJ
Jql
qlIM
'JJoJ
Jql o1 aAIIPIaJ
sarard
-peaq
eql
Jo
uortetuJrJo aqt sa8ueqr
pue
sJ)llaq
a8urq aql stdnrsrp re)npur
;o
Surpurg
'uots
-sardar
Jspelal o1 alenbape sr JerueJlJl
rossardar
aqt ot
rJJnpur
Jo
salnlelou oul
Jo
Surputg
'uta1
-ord
.rossardJJ
Jql ur
a8ueqr
IpuorlpruJoJuof,
JlerpJrurul
up sJSnp) rJ)npur
;o
Surputg
'Surpurq
HIH
JoJ a,roor8 .roleru
eqt stue
-rro
pueq
slql
'.E7-
,(q
vNq
aqr Surpurq
'yNq
:olBrado
1o
anoor8 Jourru Jql olut xqaq a8utq
uoqs
aql
Io
uoIuJSuI
aql st Sutputq
Jo
tuJuela
.{a1
y
'rolerado
aqt ro;.dlrur1;e saseanut
Llsnoru
-roua
srqJ
'anoor8
roferu
aqr
Jo
suJnl enrssJ)
-Jns
olur 3u1lrasur
IqVXq
peluoJ
tlun
Jrreurp
p
ur
surpruop Surpurq-y1qq o,lrl Jql
lpql
sMoqs
d
t'fr1
jjtitltli:i
'Jtrs-Jleq
auo ol Surpurq
qrea ,{1sno
-Juellnturs.roterado
Jqt
peluo)
o1
sarardpeaq
o,l.rl sMollp rossardar
Dplur
Jo
ruroJ JrrJrurp
Jql
leql
sr J)uJrJJJrp eqt JoJ uospJr aq1
'rossa;dar
tJelur
Jo
leql
upql ssel apnlru8eru
Jo
sJapJo
.{ueru sr TaAeMoq
'vNq1
roJ Llrurge sl1
':ossardar
]f,elur
sP alrs-JlPq e
qlrm
sl)eluo)
;o
uraued
arups
Jql Surleru [q
VNO
roterado 01
purq
up]
U
'eJoJ
eql
Jo
luapuadapur
dla,ruelar
sr aratdpeaq
eqt
qJrqM
ur
lJporu
e
pa1sa83ns
1;o,ra
,{1:eg
'sllPlu0l
snoauPllnurs aleur
o1
frlauoa6
lq6g
aql u!
lou
orp salLs burpuLq
-VN0
oMl aql
lpql
os uorleuuoJuor aq1 sebueqr
pup xrtaq
a6utq aq1 sldnrsrp 6urpurq ralnpul
r
'xrlaq
alqn0p
oql
J0
surnl a^rssellns
olur
ilasu!
upl laurp
p
Jo
surpurop 6urpuLq-yp6 otvrl
aql
qlrqM
ur u0rleur0]uor e sPq lossalOo.l OA|.|J!
o
'vN0
roleraoo
1o
enoor6 rourur eql
olur silosur
xLlaq e6uLq eq1
.
'VN0
Jo
anoo.rE rolpru
oql olur. slasur
]aurp
p
urqlrM
lauouour
qlpa jo
ureuop 6urpurQ-VN0
eLll
.
uor.lPruroJuol ur
abueqS lualsollv ue
Aq
'VN6
ol 6uLputq
sltut
-rad
lpql
uorlpluauo
ue
u1 rabuol ou
elP louttp rossaldal
1o
suoLbar
HIH
oql
pue
sploJun
xttaq abutq aql
leql
os
arol aqlJo
arnllnrls
aq1 sabueqr
lelnpul
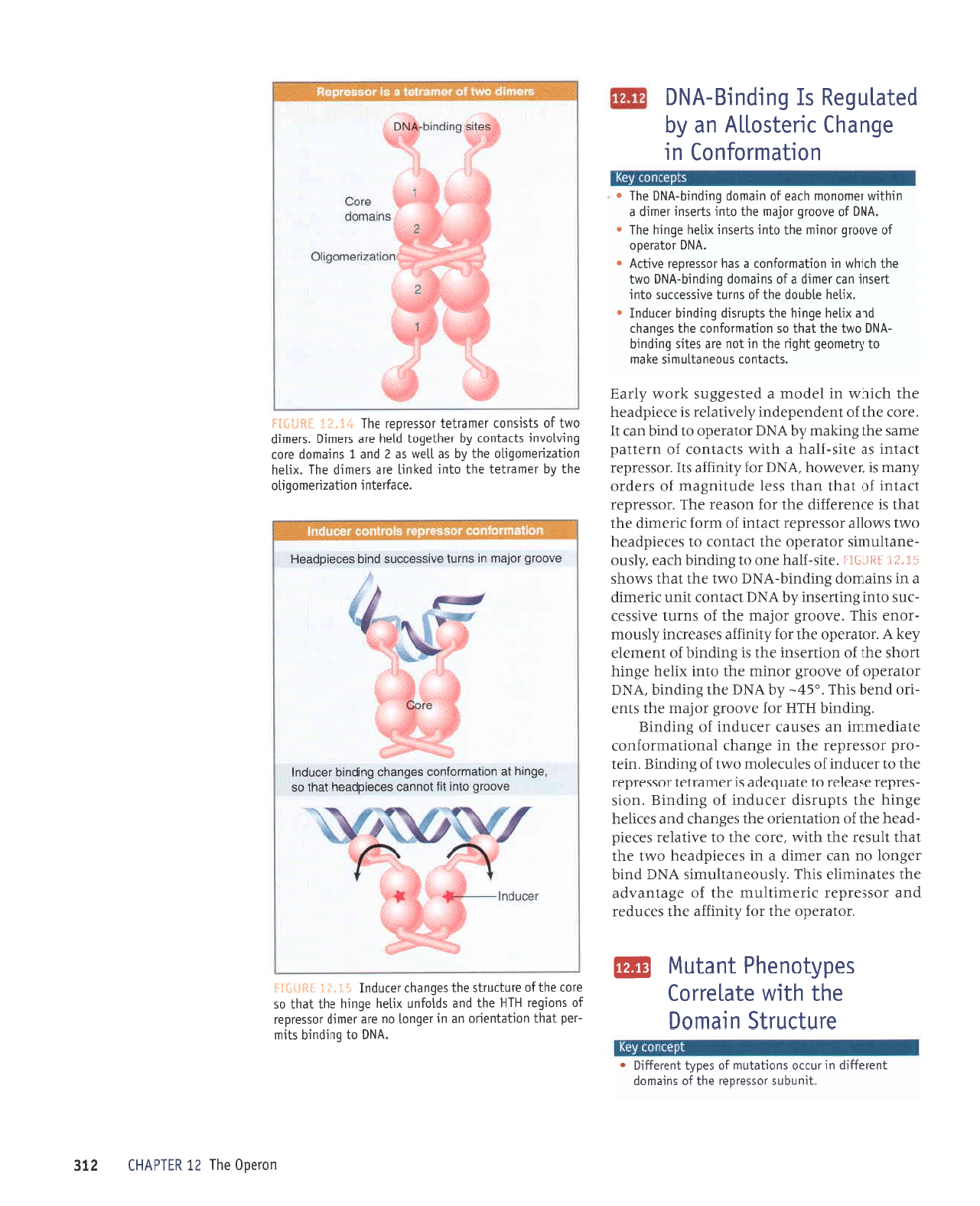
GI"i I
lS{lt3:l
anoorb olur
irl louuec
socsrdppor.l
leql
os
'e6urq
1e
uorleuroluoc
sa6ueqc 6utputq
tecnpu;
anoorb roleru ur
surnl onrssoccns
putq
sacatdp€oH
'alPJrelu
I uorlezuauoDtlo
eq1
r\q
ratuerlal
eql olut
palutl
a.lp stautp
eqt
'xtlaq
uoLlezuauobLlo
aq1 [q se
lleM
sp
Z
pue
I
suteuop oJo]
burnlonur
sllpluol
Aq raq1e6o1
ploq
arP
sleut0
'sJautp
oMl
J0
slstsuor
raue.llal rossa.loa.l
aql
tE'Ei =sfi*I*
uorao0
aql
zI
ultdvHl
zlE
pelPln6au
q
6ulpu!B-vN0
Mutations
in the
Lac repressor
identified
the
existence
of different
domains
even
before
the
structure
was
known.
We
can now
explain
the
nature
of the mutations
more
fully
by reference
to
the structure,
as
summarized
in
,
lr,l:':r
: L
.,.
Recessive
mutations
of the
lacl-
Iype
can
occur
anl.where
in the
bulk
of the
protein.
Basi-
cally, any mutation
that inactivates
the
protein
will have
this
phenotype.
The
more
detailed
mapping
of mutations
on to the
crystal
struc-
ture in
Figure 12.13
identifies
specific
impair-
ments
for
some of
these mutations,
for example,
those
that affect
oligomerization.
The
special
class of
dominant-negalive
lacla
mutations
lie
in the
DNA-binding
site
of the
repressor
subunit
(see
Section 12.9,
Multimeric
Proteins
Have
Special
Genetic
Properties).
This
explains their
ability
to
prevent
mixed
tetramers
from
binding to the
operator;
a reduction
in
the
number
of
binding
sites reduces
the specific
affinity for
the operator.
The
role
of the
N-ter-
minal
region
in specifically
binding
DNA is
shown
also by its location
as the
site of
occur-
rence
of
"tight
binding"
mutations.
These
increase
the
affinity
of the
repressor
for
the
operator,
sometimes
so much
that it
cannot
be
released
by inducer.
They
are rare.
Uninducible
/acls
mutations
map largely
in
a
region
of the
core
domain I
extending
from
the inducer-binding
site to
the hinge.
One
group
Iies in
amino acids
that
contact
the inducer,
and
these mutations
function
by
preventing
bind-
ing
of inducer.
The remaining
mutations
lie at
sites that
must be involved
in transmitting
the
allosteric
change in
conformation
to the hinge
when inducer
binds.
Repressor
Protein
Binds
to the
Operator
o
Repressor
protein
binds to
the doubte-stranded
DNA
sequence of
the operator.
r
The
operator is
a
palindromic
sequence
of 26
bp.
o
Each inverted repeat
of the operator
binds
to the
DNA-binding
site of one repressor
subunit.
The repressor
was isolated
originally
by
purify-
ing the
component
able to
bind the
gratuitous
inducer
IPTG.
(The
amount
of repressor
in
the
cell is so small
that in
order to
obtain enough
material, it
was necessary
to
use a
promoter-
up mutation
to increase
lacl
Iranscription
and
to
place
this /acllocus
on
a DNA molecule
pres-
'
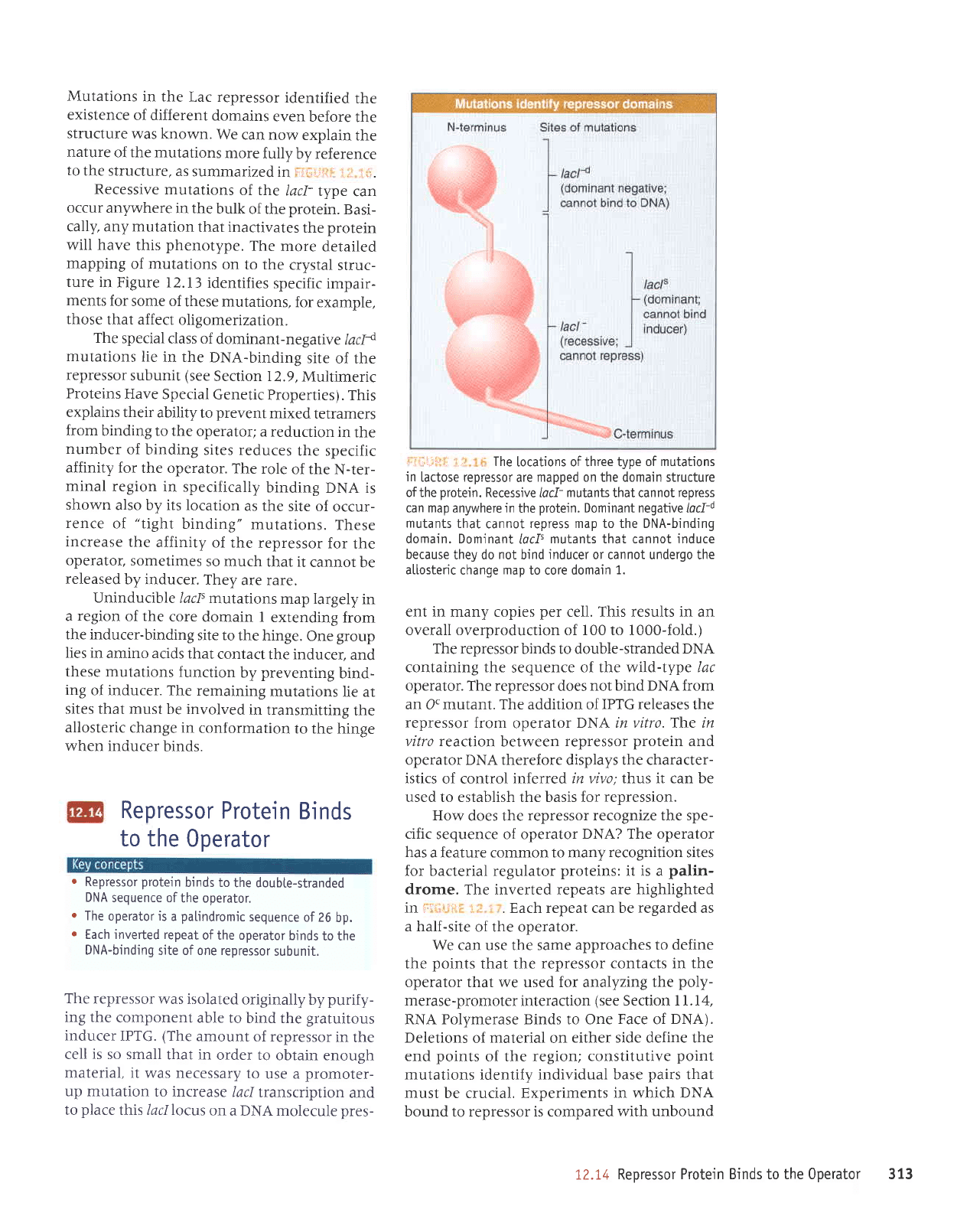
,l,l-li::, 1,,
The
[ocations of three type
of mutations
in
lactose repressor
are
mapped on the domain structure
ofthe
protein.
Recessive
lacf-
mutants
that cannot
repress
can map anywhere in
the
protein.
Dominant negative lacli
mutants
that
cannot
repress map to the DNA-binding
domain. Dominant
locf'
mutants that cannot induce
because
they do
not
bind
inducer
or cannot undergo the
allosteric
change
map
to core domain
1.
ent in many copies
per
cell. This results in an
overall overproduction of 100 to 1000-fold.)
The repressor
binds to double-stranded
DNA
containing the
sequence
of the wild-type lac
operator. The repressor
does
not bind DNA from
an OC mutant. The addition of IPTG
releases
the
repressor
from
operator
DNA in vitro. The in
vitro reacion
between
repressor
protein
and
operator DNA therefore displays the character-
istics
of control inferred
in vivo;thus it can be
used to establish the basis for
repression.
How
does the
repressor recognize the spe-
cific
sequence of operator
DNA? The operator
has
a feature common to many
recognition
sites
for bacterial regulator
proteins:
it is a
palin-
drome. The inverted repeats are
highlighted
in
i
ri
ri
r , . Each repeat can be
regarded
as
a half-site
of the operator.
We can use the same
approaches to define
the
points
that the repressor
contacts in the
operator that we used for analyzing the
poly-
merase-promoter
interaction
(see
Section I l. 14,
RNA Polymerase Binds to One
Face
of
DNA).
Deletions
of
material
on
either side define the
end
points
of the
region; constitutive
point
mutations
identify individual
base
pairs
that
must
be crucial. Experiments
in which DNA
bound to repressor is compared with
unbound
12.14 Reoressor
Protein Binds to the 0perator 313
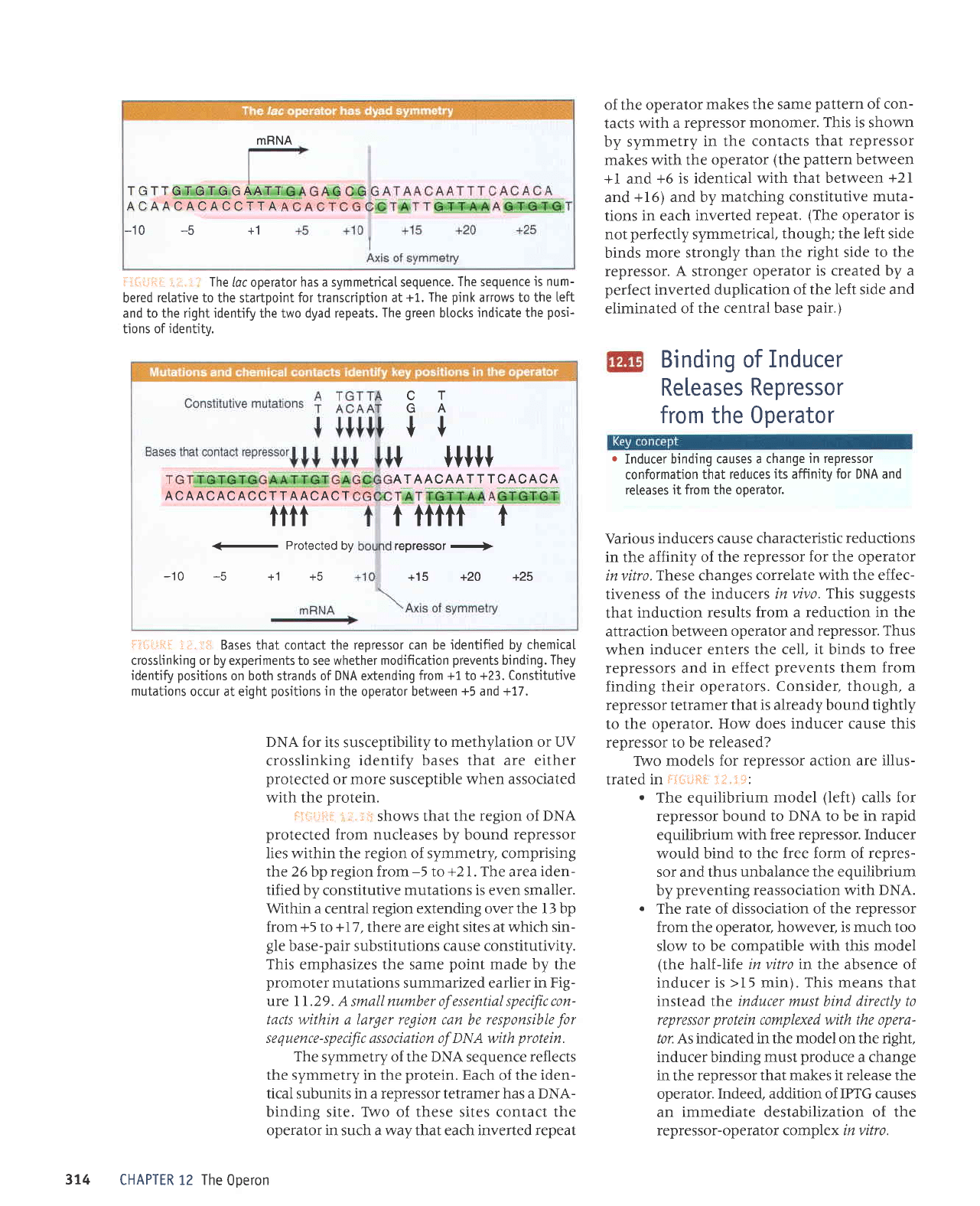
MRNA
TGTTGTGTG
GAATT
ACAACACACCTTA
-10 -5
+1
of the operator
makes
the same
pattern
of
con-
tacts with
a repressor
monomer. This is shown
by symmetry
in the contacts
that repressor
makes with the operator
(the pattern
between
+l and
+6 is identical with
that between
+21
and
+16) and by
matching constitutive
muta-
tions in each
inverted
repeat.
(The
operator
is
not
perfectly
symmetrical,
though; the left side
binds
more strongly
than the
right
side
to the
repressor.
A
stronger
operator
is created by a
perfect
inverted duplication
of the left side and
eliminated
of
the central base
pair.)
iii:r.ji.:i
:.j. i
.r
The
loc operator
has a symmetricaI sequence.
The sequence
is num-
bered
retative to
the startpoint
for transcription at
+L, The
pink
arrows to the
teft
and to the right identify the two dyad repeats.
The
green
blocks
indjcate the
posi-
tions of
identity.
i:liri.ii1li.
-i,i"
l:;ri Bases that
contact
the repressor can be identified bv
chemicaI
cross[inking or by experiments to see whether modification
prevents
binding.
They
identify
positions
on both strands of DNA extending
from +1
to
+23. Constitutive
mutations
occur at eight
positions
in
the operator between
+5
and +17.
DNA for its
susceptibility
to methylation or UV
crosslinking
identify bases that are either
protected
or more susceptible
when associated
with the
protein.
iii".iiiii:
::j.:ii
shows that the
region of DNA
protected
from nucleases
by bound
repressor
lies within the region of symmetry, comprising
the 26 bp region from
-5
to
+2
l. The area
iden-
tified
by constitutive
mutations is even smaller.
Within a
central
region extending over the l3 bp
from +5
Io
+I7
,
there are eight sites at
which sin-
+[iffi
Jffi::1T:"':nfi
,ff ni:Htx
promoter
mutations
summarized earlier
in Fig-
ure I I.29. A
small
number of essential specific con-
tacts within
a larger
region
can be
responsible
for
sequence-specific association of DNA with
protein
The
symmetry of the DNA sequence reflects
the symmetry in the
protein.
Each
of
the iden-
tical subunits in a repressor tetramer has a DNA-
binding site. TWo of these sites contact the
operator in such a way that each inverted repeat
CHAPTER 12 The
0oeron
CT
GA
t+
ACAACACACCT T AACACT
tttt
-
Protected
by
-10 -5
+1 +5
+
lltt+
AJAAc_44If
rc49494
t tfttf t
repressor
+
+15 +2O +25
Binding of
Inducer
Releases
Repressor
from
the
0perator
r
Inducer binding
causes a change
in repressor
conformation
that reduces
its affinity for DNA anc
releases it from the operator.
Various
inducers cause characteristic
reductions
in the affinity of
the repressor
for
the operator
invitro.
These changes
correlate with the effec-
tiveness of
the inducers in
vivo. This
suggests
that
induction results
from a reduction in the
attraction between
operator and
repressor. Thus
when inducer
enters the cell,
it
binds to
free
repressors and
in effect
prevents
them
from
finding their operators.
Consider, though, a
repressor tetramer
that is already bound tightly
to the operator.
How does
inducer cause this
repressor to be
released?
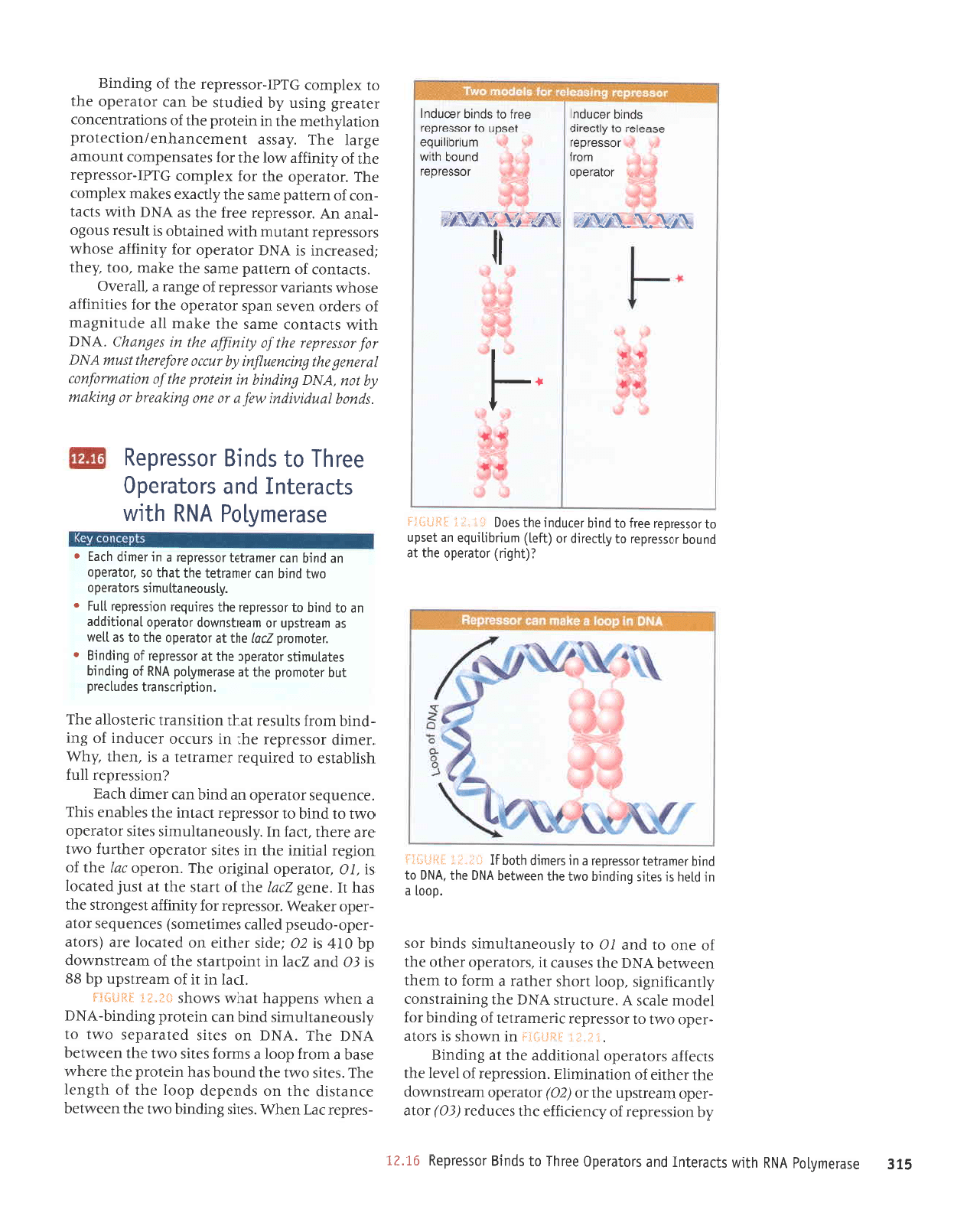
TWo models
for renressor action are illus-
trated
in
FI"";iisi.
:.;:.:*s:
.
The equilibrium
model
(left)
calls for
repressor bound to
DNA
to be
in rapid
equilibrium with
free repressor. Inducer
would bind
to the free form of repres-
sor and thus
unbalance the equilibrium
by
preventing
reassociation with DNA.
.
The rate of dissociation of
the repressor
from
the operator,
however, is much too
slow
to be compatible with this model
(the
half-life in vitro in the absence of
inducer is
>I5
min). This means that
instead the inducer must bind directly to
represslr
protein
complexed with
the
opera-
/or As indicated in the model on the right,
inducer binding must
produce
a change
in the
repressor that makes it release the
operator.
Indeed, addition of IPTG causes
an
immediate destabilization of the
repressor-operator complex in vitro.
314
9IE
asprau^lod
VNU
qllM
sllpletul
pup
sroleledo
aalql
ol sputg rossaldau
9I.ZI
Lq
uorssardar yo
druanr;ye
aql srrnpa
t
(gg)
rc1e
-rado
rueapsdn
aql ro
(79)
rcpndo
rueJllsuuop
Jqt JJqtrJ
Jo
uorteururrlE
'uorssardar
Jo Ie^JI
Jql
stJJlJp
srolerado
Ipuoutppe
aql
te
Surpurg
':l':
I
iEfi:_::j
uI
uMoqs
sI sJole
-rado
ozral ol
rossardar
JrJerupJlal;o
3urpurq ro;
IJporu
elp)s
V
'aJnDnJts
VNe
eqt
Sururertsuot
,(11uerr;ru8rs
'doo1
uoqs
Jeqtel
e ruJoJ
ol rueql
ueaulJq
VNo
Jql sasneJ
1r
'sroterado
Jaqlo
aql
Jo
Juo
ot
pup
IO
ot
^lsnoJuellnurs
spurq Jos
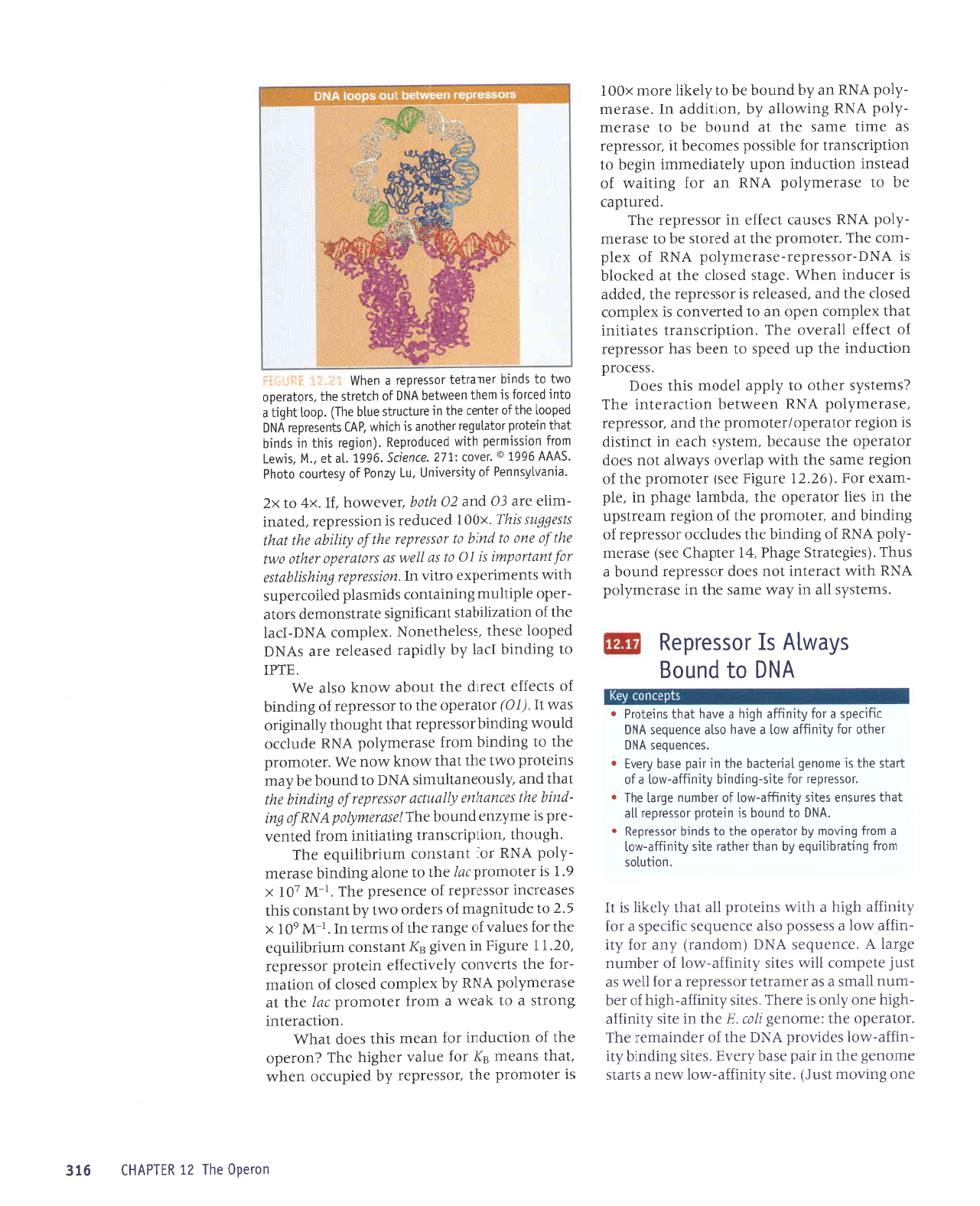
'd00]
P
ur
plaq
sr
selrs 6urpurq
oml aql
uaaMlaq
VNq
eql
,VN6
ol
purq
.rauetlal
lossatdat
e ur slaurp qloq
lI
*i:'{: i
:l:J* -ill
iGqEy)
rolerado
aq1
1e
punoq
rossetdat
o1 fi11rerLp
to (ga1)
urnuqrtrnba
ue
lasdn
o1rosseldet
oal1
ol
purqlolnputoqlseo0
+i.,:i
]Sal*i-j
-sardar
)pf uaq^A'sa1rs
Surpurq
o,lrl eql
uaamleq
eJuptsrp
Jqt uo spuadap
dool
aqt
yo
qt8ual
aqJ'selrs
oml eqt
punoq
seq
uratord
eqt eJeqM
JSeq
e ruor; dool
e suroJ
sJlrs oml
eql uJaMlJq
VN61
JqI
'VNq1
uo sJlrs
pJtprpdas
o,ul
ol
,{.1snoaue11nurs purq
uer
uralord
3urpurq-y51q
p
uJqM suaddeq
teqm
suoqs
*I"frE
3Hf:*1.,4
'IJel
uI
l1
;o
ruea;1sdn
dq
gg
sr
tO
pue
ZJel
ut
luroduets
aq1
Jo
ueeJtsuMop
dq
Otl
sr
ZO
:aprs
rJqtrJ
uo
petp)ol
are
(sro1e
-rado-opnasd
palpr
saurrlaruos)
saruanbas
role
-rado
ra1ea14
'rossatdar
roJ
^tlulJJp
lsa8uorls
aql
seq
11
'aua3
nq
JI,trLJo
uels
eql
]e
lsnf
pJleJol
sr'/O
Toteredo
pul3rro
aq1
'uorado
)q
eqtlo
uor8ar
Ipnrul
aql ur
salrs roterado
JJqunJ
o,lrl
eJe aJJql
'peJ
uI
'dlsnoauellnurrs
sJlrs
roterado
ol!\l
01
purq
o1 rossardar
lJplur
aql sJlq€ue
srqJ
'aruanbas
JotpJJdo
up
purq
upl raurp
qJeg
2uorssarda:
1n;
qsrlqElsJ
ol
peJInbJJ
JaureJtJt
e sr
'uaql
z(q7y1
'Jarurp
rossardar
Jql
ur sJnlJo
JJJnpur
;o
8ur
-purq
ruoJJ sllnsaJ
leqt
uorlrsueJl
JrJelsolle
eqJ
'
uoqdursuerl
sapnlrard
1nq
ralouotd
aq1
1e
eserauflod
ypp
1o
6urpurq
selelnuqs
rolerado
eq1
1e
tossardar;o
6urpurg
r
'lalourotd
?Dl
aq11e
loleredo
aql ol
se
lla/v\
sp
ueollson l0
ueallsuMop
lolplodo
lpuotltppp
up
01
putq
o1
tossardar aq1
serrnbar
uorssaldat
11ng
.
'[lsnoau
e11 n urrs
sroleredo
oMl
purq
upl
laupllal
aql
lpql
0s lolptado
up
purq
upl leupllal
lossatdal
p
u[
]aurp
r])pl
o
asPraunlod
vNU
qt$
spPjaluJ puP
sjolPjad0
oarqI
o] spurg
tossardau
'spuzq
pnpt^tput
mal a n
auo 6ut4aatq
n 6ut4aw
lq
pu'VNe
6u1pu1q
w utalotd
atpto
uo4awotuot
Tatauaf
a4t
Qunuani[ut
[.q mno
atotata4t
$nru
VNe
tot tossatdat
aLlJt0
tjtutlla
a4l
ut sa6ua43
.yyq
qt!&t
slJeluof,
Jrups
eql
J>lpru
11e
apnlru8eur
Jo
sJapJo
uanas ueds
roterado
eql
JoJ
senrurJJp
asoqM sluerJel
Jossardar;o
aSuer
e
,llpJelo
'slJelUOJ;O
Uralled
alups Jr{l
a>leur
'oot
z(aqt
lpJspaJJur
sl
vfd(
rolerado
ro;
dlrur;;e
JSoqM
srossaldar
luplnru
qlrm pJurplqo
sr
11nsar
sno8o
-lpup
u17
'rossarda.r
earJ
Jql sp
VN11
qll,lr
sDel
-uoJ
Jo
uJallpd
Jrrrps aqt z(prexa
sa>1eru
xaldruor
aqJ
'rolerado
Jql JoJ xaldruor g141-rossardar
Jql
Jo
^lrulJJp
,lrol
eqt
roJ satesuaduroJ
lunoure
aBrel
aq1'z(essB
luaurJJupqua/uollralo.rd
uor1e1,{.qtaru
aql ur
uralord
aqt
yo
suorteJluJJuo)
ralear8
Sursn ^dq pepnls
aq
ueJ
rolerado
aql
o1 xaldruot
)JdI-JossJJdar
aql;o
Surpurg
v\\\,{\{.'
toleredo
utoll
rossarder
eseelet
o1 I;lcelp
sputq
Jocnpul
t
I
+-1
I
II
tl
\,#_4\ FLft#'
rossetdat
punoq
Llu^
unuqrllnbe
]oson
o] rossojda.l
oajl ol
spurq .tocnpul
uoredo
aql
zI
ulldvHl
glg
Juo 8ur^oru
lsnf)
'etrs
Llrur;;e-,r,ro1 Mau
p
stJpls
aruoua8 aqt
ur rred aseq.d:a,Lg
'salts
Sutputq z(tr
-urJJp-Mol
saplr.ord
vNo
eqt
Jo
rapureuJr
rqJ
'rolerado
aqt
:aruouaS
4n'E
erlt ut
alts Lltur;;e
-q8rq
auo
.dpo sr rreql'setrs,{lrugye-q8q;o
raq
-unu
llerus
e se JJurprlJl
rossardar
p
JoJ
IIJM
se
tsnl
aladuroJ
IIrM
salrs
,(rluUJB-uol
Io
JJqrrnu
a3re1
y
'aruanbas
y51q (ruopuer)
,{ue ro1 L1r
-urlle
Mol e ssassod osle aruanbas
rtytrads e ro;
.dlrurge
qBIq
p
qUM
surrlord
1p
reqr
,{.1a>1t1 st
r1
'u0qnl0s
ruoll
6urlerqrlrnba
fiq ueql leqler olts
f1Lu4e-no1
e urorl
6ur,rou
fiq
rolerado
eql ol sputq rossardaX
.
'VN0
ol
punoq
sr uralord .rossardar
11e
leql
salnsue
selrs l{ltu
;e-rvrol Jo
laqunu
e6re1 eql
r
'rosserdar
rol elLs-6utpuLq
[1Luq1e-mo1 e
1o
1.rpls
aql sL euoua6
lpuallpq
aql u! llpd aseq
,{ren3
r
'soru0nDas
vN0
.laqlo
loJ r{11ug1e
/v\ol
p
a^eq osle
aruanbes
y;r16
rgoads
e ro; [1rug;e
q61q
e aneq
]e!]
sur.o]orl
o
vNo
ol
punoB
sAelrnly
sI rossardau
'sruals.ri.s
11e
ur
,(e.tr Jues eql ur aseraurdlod
VNU
ql1n,r
t)pJetul lou
sJop
rossarda.r
punoq
e
snql'(sar8aterl5
a8eq4'71 raldeq3 aas)
ase.raru
-.{1od
y1qg
1o
Surpurq Jqt
srpnlt)o rossa.rdar;o
Surpurq
pue
'rJtoruo-rd
aql
;o
uo6ar
ruearlsdn
Jqt ul sa11
rolerado Jqt
'ppqruel
a8eqd
ur
'a1d
-rupxJ
roJ'OZ'ZI
arnSrg aas)
raloruord aqr
;o
uot3ar
erups Jql
qllrrn
depa,ro sf,e,u.1e
lou
seop
rolerado
eql asne)Jq
'urals.ds
qlpa
uI
llullslp
sr uorBar
roleradoTraloruord
eql
pue
lossarda.r
'aseraruLlod
VNU
uJaMlJq
uoltJeJJtul
eqJ
Zsruatszi.s
reqlo
ot .{1dde
lapou
slqt
seoc
'ssarord
uort)npul
aqt dn
paads
o1 uaaq ser{ lossatdar
Jo
tJJIJa
IIelaAo
aqJ
'uorldtr)suell
sJleIlIuI
leql
xalduro:
uado ue ot
pJuJ^uor
st
xaldruor
pJsop
eql
pup
'peseJleJ
sr rossardar
Jql
'pJppp
sr Jef,npur
uJqM
'a8ets
pasoll
Jql
te
pe>lJolq
sr
vNq-lossardar-aseraurLlod
y51g
yo
xald
-urol
eql
'ralouord
aql
le
pJrots
Jq 01 espJJru
-.{1od
y111g
sJSneJ
traJJJ
uI
rossa,rdar aq1
'pa;n1der
eq
o1 aseraruzllod
vNu
ue
ro1 3utlte.tr
;o
ppalsur
uorlJnpul
uodn
[lalerparurut
ut8aq o1
uorldrnsuBrl
ro; alqrssod saruoJJq
ll
?ossarder
se eurrl
Jues
Jql
le
punoq
eq o1 JseJau
-,(1od
y1qg
3ut.uo11e
Lq
'uontppe
uI
'rseretu
-,(1od
y111g
ue
z(q
punoq
aq ol.d1a41erou x00I
sr ;alotuord
aqt
'rossardar
.dq
pardnrlo uJqM
'tpqt
supJru
s)1
roJ
anlen
raq8rq
aq1
;uorado
Jql
Jo
uolDnpul
JoJ
uPau
slql
seop
lptl^A
'uoIlJeJalUI
Suorls
e ol
>leJM e
rrroJJ
taloruord
)al eqt
w
aserau.{1od
VNU
^q
xalduror
pesolJ
Jo
uolleru
-rol
Jql suJluo)
Llaa,rna;;a
utalord
rossaldal
'g6'1
1
a.rnBrl
ut uantB
B)/
luelsuoJ
urntrqrltnba
Jqt JoJ sanlel
Jo
a8uer
aq1
Jo
suJel
uI
'r-W
o0I
x
S'Z
ot
epntlu8eru
;o
srepJo
o,trt
,(q
luetsuoJ
slql
sJsparJul
rossardar;o
aruasard
aI{J'r-W
r0T
x
6' I
sI Jelouo
td n1
arTt 01
auole
Sutputq
eseJJrrr
-L1od
y51g
roJ
tuPlsuo)
unr.rqtltnba
aq1
'q8noql'uotldtllsuell
Sutletltul
luoJJ
pJluel
-a.rd
sr au^,izuJ
punoq aqTiasataw[1ody1t1ylo
6ut
-pwq
aLli snua7ua
[11anpa
nssatdat
to
Cutputq
a4l
leq]
pue ,{lsnoauelpruls
YNC
ot
punoq
aq
zieru
suralord
o.l.tl Jql
tPqt
,trou>I
Mou
3M'raloruord
rqt 01 Surpurq
urorJ
asPrJur,{lod
VNU
rpnlJ)o
plno.tt
Surputq
rossarda:
teql
lq8noqt
^dlleur8uo
selv':4'(td
rolerado
Jqt
ol
rossJrda.r;o
Sutputq
Jo
sllJJJe
lJaJIp
Jql
lnoqe
Mou>l
osle
a^A
.fIdI
or Surpurq
1re1
zlq
.{.1ptde;
pasPalar ere sVN(
padool
esJqt
'sselrqteuoN
'xaldruor
VNq-IJel
eql
Jo
uoIlezlIIqpls
luP)IJIu8rs
alerlsuoruJp
srole
-rado
a1drl1nru
SututBtuor
spnuseld
paltorradns
qlrM slueurradxa
orltn
u1'
u o
ts s a t da
t 6utr4sq
qa|s
a
tot
luultodu,tt
s!
to
0i
sa
lla/A
sa stolatado
taLlp
0/w
a4l
to
auo 01
pryq oi nssatdat
a4l
lo
firy1qa
a41
ru41
s1sa66ns
sn41'x00
I
pJJnpar st
uotssardar
'paleul
-rurl3
JJP
to
pue
79
t'110Q
TJAJMOq
'II'xL
olxz
'eLuen1fisuue4
1o
filtstenrul
'n1
Azuo6
1o
fiselnor
o1oq6
'svvv
9661
6
'1oAo3
ilLZ'a2uaos'9661
'le
la
"l/ll
'smal
uor1 uotsstulad
qluvr
pernpordeg
'(uotbal
stqt ut
sputq
1eq1
uralord
rolelnbar
raqloue
sr
qltqm'dVl sluosaldal
VN6
pedool
aq1
1o
ralual
eqt ur
e.rnpnils
enlq
aql)
'doo1
lqbq
e
olur
pal.lo; st uiaql
ueamlaq
VN0
Jo
qllalts eq1'stolelado
oMl ol
sputq .leuP.llal
losseldai
e uoqM
L.
.'! rrriil-li:

LIE
lossardau pu!g
ol sa]ts flruq.lv-Mol qlrM
saladuol rolprado
aql
gI.ZI
Jqt
tpql
arnsuJ
srqt seop ,lroH
'etrs
(ruopuer)
.{UugJe-rvrol
tlue
ueql JJuaq
16l
rossardar
aql
ro1 aladruor
IIrM
lpqt
alrs
dlrury;e-qErq
afurs e
sasrrdruor
rolerado Jqr
snqJ
'qfual
atups aql
Jo
aruanbas
yNC
ruopuer z(ue
ot ueql
VNO
Jole
-;ado
ol JJllaq sJtult
z0I-
spurq rossardag
'rotBrado
Jql
ruoJJ alerJossrp
ol
lr
sesnpJ JaJnpur
uJqM
rossardar
aqt o1 suaddeq
lpqm
pup yNq1
Jo
lsal
eql
pup
,roterado
Jql uJJMleq
pJuollp
-red
sr
rossardar Moq
J)npap up)
a^lur
'sluplsuoJ
asaql uor{
'Surpurq
VN(
IpJeuJSTrossardar
qllzvr
Sutputq Joleredo/Jossardar
rrl
JoJ slupls
-uof,
rxnrJqrlmba
aq1 sareduror
Ee'eI
lupSIJ
'slaBrel
ra,nay
qlrM
sroteln8ar
ueqt
saltpuenb ralear8
ur
punoJ
aq
ol sta8ret Lupru
qlru
sJolep
-3a:
Dadxa
a,lr os
'sJlrs
ta8rel
rr;nads;o
Jeqrunu
Ie1ol
aql
Jo
ssJJXe
JlqpuospaJ
uI eq
lsnru
oslB uralord
Jo
lunotue
"qI
.
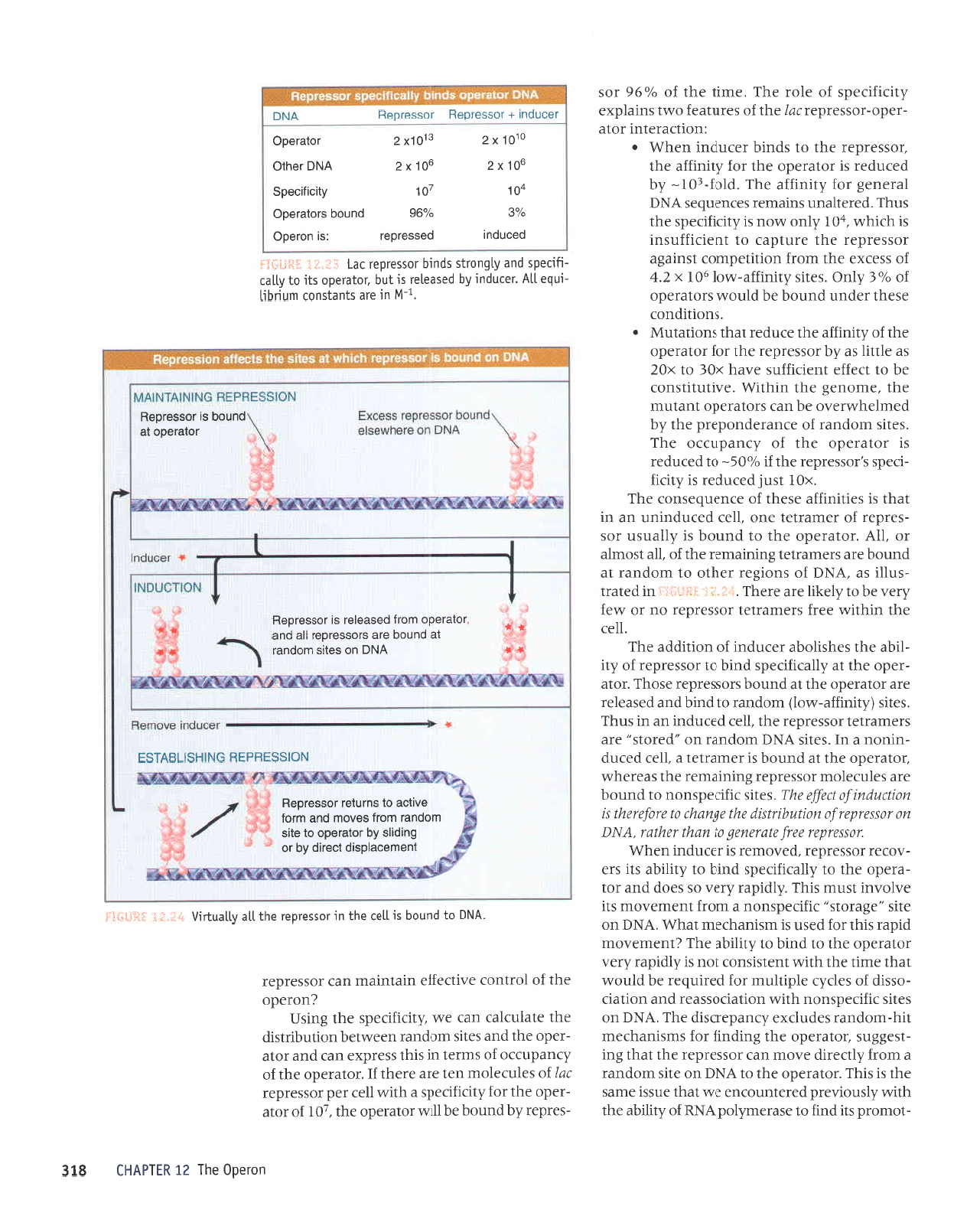
'^lPIJIJEOS
Jqt
qlrm
sJspaJJep pue
aruoua8
Jql ur
VN(
JO
1UNOIUP
Ie]Ol
Aql
qlIM
SJSEAJJUI
parrnbar
sr
tpqt
ulatord
Jo
tunotup
ar1J .
'YNC
Jo
sselx
eql
Jo
DeJJr
rqr
sralunof,
uralord
aql;o
,hnr;nads
aq1 ,
'se1rs
la8rel
rr;nads
pulq
01 uratord
e
;o
,{1r
-llqp
Jqr
setnltp aruoua8
aq1
Io
azrs e{J .
:are
sraletueted
lueuodtul
aql ,{1anrlrn1ur
papadxa
aq
lq8nu
sy
'Surpurq
rr;oadsuou
pue
rr;nads JoJ suorl
-enba
runrrqqrnba
aqt Surredruot
[q atrs
la3rel
slr etpJntes
o1 uralord
ro1e1n3ar
e
yo
[1rpqe
aql
eJuenllur
lpqt
srJleruered
aql aurJJp
upJ e711
'oAu
uL
vNo
Jo
uoqellualuol
a^qla#e
aql ur uorllnpat
lo
aspailur ue Aq
pabueqr
aq
plnol
slalatuered
aseql
.
'luaualel0srp
palrp
[q alrs rlruu-le-ivro1
e o1 rolerado
aql uol]
a^out ol lossaldal
sosnpJ
uorl]npu[
o
'punoq
ere
srolerado
;o
gog
Aluo
leql
os
'sa1s
Alrug;e-rvro1
Jo
lpql
xr6l
o1 rolerado
aq1 rol flrugle
eql
salnpal uor]lnpul
o
'autl
aql
Jo
o/096
lossaldal r{q
punoq
sr loletado
aql
leq}
selnsua
11ar
rad
staupllal lossardat
ual
lo la^al
eql
.
'elrs
Alrul#P
Mol
s
J0
leql
x10I
sr
1eq1
lossardet
ro; AlLugle
ue seq
rolerado
aql
'latnput
Jo
otuesqe
a{} ul
r
lossaloeu pu!g
ot
satls Allu!#v-Mol
qtm
'drlrlJlrads
aqt
'sr
1eql
'(aJuenbas
ygq
ruopuer
z(ue
Surpurq roJ
tuelsuo)
aql)
d'51
ol
(atrs
rqneds
e Surpurq
JoJ
tuplsuoJ
Jql)
ds)
Jo
onpr eq]
sl
tueuodru
sI
lpq,vt.
llupuodrul
tou
Jre
vNO
ruop
-upJ
pue
rolerado JoJ stuplsuoJ
uorlerJosse
eql
Jo
sJnlpA Jlnlosqp aqt
'uortrtadruof
srql uI
'selrs
AlrurJJP-1!l.ol
Jo
Jeqrunu
a8re1 aql
ql\m
sapdwoc rolerado
aql
;o
atls
zhtug;e-q8q
al8urs eqt
rprqM ur
'vN(I
uo ros
-sa:da.r
aql
1o
Cutuotqtlta d Jql r+IM
pauJJJuoJ
JJe
aM
lpql
sueeur
1I
:JolpJado
aql
qllM
rossardar
Jo
uorlJpJatul
eql JoJ uorlerr1durr
lueilodrur
ue
seq srqJ
'uralord
rossardar aaJJ
ou sr eJJql
tpql
Surdes 01
lunoruelupt
sl slqr
1ar
rad rossardar
Jo
selnJJloru
0I-
eJp JrJql
e)urs
'VNA
(wlp
-uat)
ol
punoq
s nssatdatto
o/o
t0'0
fiq
lp
snLLI
'r0I
=
rossardar
punogi
eeJd
:sa,tr8
sanle,r
asaql Suuntusqn5
'(uorlerluaJ
-uor
q8q
zhan
e)
W
s-0I
x
Z
=
[VXq]
o1 spuodsarrol
q)rr{M
'JaUI
sr_0
I
Jo
arunlol
IprJaDpq
p
ul
s0I
x
7
s1 salrs Eur
-purq
rqnadsuou
Jo
uorlpJluJJuo)
aef o
'
1-W e0I
x
Z
=
v)I
sl
luelsuoJ
Surpurq runrrqrlnba
trynadsuou
aVJ .
:]eql
purJ
aM
'tna1strsw..
Jql JoJ sralaruered
aql Surdlddy
'JOSSJJCIeT
JaJJ
JO
UOU
-rodord
aqt
a,rr3 o1
pa8uerreal
aq upJ rossardar
punoq-vN(
pue
rossardal
aeJJ uJJMlJq
runrJqrl
-rnba
aqt SurqrJJsep
Jo
uopenba
Jqt zlroq
suoqs
u e'eI 3ufiglj
'uorlnlos
ur aaJJ surprual
euou
pup
VN(
01
punoq
sr
rossardar
1e
Llpngm
ro
ilp
'alrs
Supurq rgoads
p
Jo
JJuasqE
Jqt
q
uele
'leq]
sueaur salrs zhrugJe-,lrol
Jo
Jaqunu
a8rel aq1
'SJTIS
AIIUIJJE-MOI
90
I
X
Z'V
Jrc
EJAI{I SNqJ
(iarls
drlulJJe-Mol
e serpeJJ
'Jlesrl
JoteJado
aqr
qryr,r
aseqd
Jo
lno
'aruoua8
aqr
Suop rred
aseq
'uorlenba
unuqrlrnba
ue {q
peutano6
sr salLs
uropupr
o1 6urpurq tosserdag
ZZ.aT
iEfiglj
lVruOl
x
v)
[y16-rosserdeg]
t
=
losserdar
earSl
:uorlenbe
eq1 6urbueleel
Iq
uenrb
sr rosseldet eerl;o
uoryodotd
eq1
IVwOI
[rossarder
earl]
JvNa-rossaldeHl
=')
:uorlenba
aq1 Iq
pequcsep
sl
VNCI
(ruopuer)
o1 Ourpurq rosseldel
rol
unuqrlrnbe
eq1
seladuol
rolPtod0
eql
-loruord
s1I
puIJ
01 eserJru^lod
VNU
Jo
,{11gqe
aqt
qlltr.dlsnomard paraluno)ua
e,ll.
leql
Jnssr Jrues
eqt sr srqJ
'rolerado
Jqt ot
vNC
uo etrs ruopueJ
e ruoJJ
l.1narp
Jloru ue):ossardar Jq1
lpql
8ur
-lsa33ns
tolerado aqt Surpury
roJ srrrsrueq)Jrrr
llq-uropueJ
sepnlrxa l:uedansrp ar{J'vNe uo
salls rgoadsuou
qlrM
uoll€lJosseer
pup
uorlprJ
-ossrp
Jo
salr.dr aldrtlnru roy
parrnbar
Jq
plnoM
teqt
arull Jql
qlr.tl
luatsrsuo) tou
sr
dlprder Lrarr
rolBrado eqt ot
purq
ot,{trpqe JqJ
Ztuarualou
prder
srql roJ
pesn
sr usrueq)Jru
teqM
'VNO
uo
alrs
,,a8erols,,
rr;nadsuou
p
ruoJJ
luJruenoru
slr
eAIoAur
lsnu
sn{J
'L1prder,{.ran
os saop
pue
rot
-erado
aqt ol ,{1err;nads
purq
o1
,{tr1qe
st1 sra
-Ao)JJ
rossardar
'pJAorrrJJ
sr JeJnpur uaq6
tossatdat aatt alatauaF ol uayl
JaLlwt'VNe
uo
nssatdatto
ulqnEustp
a4l aCuary o1 atotata4l st
uo4tnputto
ltalla
atfi'sa1rs rr;oadsuou
ol
punoq
eJp sJInJaloru
rossardar
Surureruar eql seaJaqm
'roterado
aql
lp
punoq
sr raureJlat e
'llJJ
peJnp
-uruou
e uI'sJlls
VNCI
ruopueJ uo
,,peJols,,
Jre
sJJrueJlel rossardar Jql
'lleJ
pJrnpu
ue
ur snqJ
'sa1rs
(,furugye-,uo1)
ruopupJ ot
purq
pue
pesee1er
are rolerado aql
1e
punoq
srossa.rdar
asoql'rolp
-rado
aqt
1e
.,i.11erryoads
pulq
ot rossardar
1o
,{.1r
-lrqe
er{r saqslloqe JaJnpur
Jo
uorlrppe JqJ
.IIJJ
aqt ulqtlm earJ
srJrueJtat
rossa.rda.r ou Jo ,tnaJ
Lran aq ot .{1a>1q JJe aJaqJ'
1r',.
i
t,
ri
rj i
:
:} ir:
ur
pelPrl
-snllr
sp
'VN11
Io
suorSar JJqlo 01 ruopueJ
1e
punoq
JJe srJureJlJl Surureruar aqt
Jo
'lle
tsorule
ro
'llv 'roterado
aqt ot
punoq
sr
Lllensn ros
-sarda,r
Jo
JJruprlal auo
'lleJ
pe)npurun
ue ur
lpql
sr sJrlrurJJp JsJqt
Jo
aruanbasuo; aq1
'xg
1
tsnI
pJJnpJr
sr
,{lr:r1
-rrads
s,rossardar aqr;1
ohOS-
ot
pa)npJJ
sr role:ado Jql
Jo
druednrro
aq1
'seus
uropueJ
Jo
eJueJJpuodard aqr.dq
pJr.ulJqMJJ^o
Jq upJ srolpJJdo
luplnur
Jqt'JruouJS aqt ulqtlM'anrlnlrtsuo)
Jq ol
DeIJr lurrrrJlns
J^eq
x0€
o1
xoz
se JI11ll se
.{q
rossarder aq1 roJ rolerado
aqr;o ,,trruq;e Jq1 JJnper
teet
suouelnW o
'suorlrpuoJ
asJql JJpun
punoq
eq
plnoM
srole.rado
lo
"/o€
^dpg
'sa1rs
Lrrur;;e-,lro1
s1I
x
Z'b
Jo
ssJJXJ Jql ruoJJ uorlrladruor
lsure8e
rossardar aql arnlder ol
luJrJrJJnsur
sr
q)rqM
'r0I
.{1uo ,lrou sr,{lnryrads aql
snr.{J'pJJJtlpun
sureurJr sa;uanbas
ypq
leraua8
ro;
.{lrur;;e
aql
'p1oy-161-
.{q
parnpal
sr rolerado eql JoJ .{trur;ye aql
'rossardar
Jql ol spurq JJlnpur uJe,\,\ o
:uorlrprJlur Jole
-rado-.rossar
dat tal Jql
Jo
sJJnleeJ oml sureldxa
.{lrrryrrads
Jo
JIor
rqJ
'arrlt
Jqt
Jo
oh96
ros
uoradg
aql
ZI
UlldVHl
-sardar
.dq
punoq
eq
IIrM
rolerado
aql
'z0I
Jo
role
-rado
aql ro;
,(tlrulrads
e
qlIM
1al
-rad rossa-rdar
Jrl
Jo
sJlnJalou
uat
aJp JlJqt;1
'roterado
aqt
yo
,{.ruednrro
Jo
sIuJJl
uI slql
ssardxa
ueJ
pue
JolP
-rado
aql
pup
selrs uopuer
uJJMleq
uopnqlJrslp
eqt JlplnJIeJ ue)
e,rt
,{1trt;nads
aql Sursn
auorado
Jqt
Jo
loJluor
eAIDJJJJ
ulelulelu
uel tossardal
'r_fl
ur 0lP sluelsu0l
ulnuqtl
-Lnba
11y
'.lalnpur
Aq
pesealat st
lnq
lolPlodo
s1t o1
Aler
-grreds
pue
A16uot1s sputq
losseldol
lPl
i:.;:'i:it ,lHil:i.i:i
8I€
'vN0
o1
punoq
sr
llol
aql
ut rossardar
aq1
1e
Alenltn
rr.'il'li lllill:.tI:l
luewaceldsrp
lcattp,{q
to
bulplls
{q
roleredo ol elts
uropueJ uroll
so^oul
pue
LUJoI
a^rlce
ol surnlor
rossoJdeu
NOtSSSUdf
U 9NlHSI-lSVrS:l
*
*
vNc
uo sells
ulopuel
- le
punoq
ere
stossaldel
1;e
Pue
+
+
)o1etedor'uorl
pesee1",
sl lossetdeg
rolelooo
le
punoq
sr Josseroou
pacnpur
possoldol :sl uoJooo
ToE
%96
punoq
slolelooo
r0
!
LOI
^ltctltcedg
eolxz
eolxz
vNcleqto
or0[xz
sroLXZ
lo]elaoo
Jocnput
+
rosserdag
rossordou
VNC

ers
(see
Figure 11.22
and Figure I1.23).
The
same solution
is likely:
movement could
be
accomplished by reducing the
dimensionality
of the search by sliding along DNA
or by direct
displacement from site
to site
(as
indicated in Fig-
ure 12.241. A displacement reaction might
be
aided by the
presence
of more binding sites
per
tetramer
(four)
than are actually needed
to con-
tact DNA at any one time
(two).
The
parameters
involved
in finding a high-
affinity operator in the face
of competition from
many low-affinity sites
pose
a dilemma for
repressor. Under conditions
of
repression,
there
must
be
high
specificity
for
the operator. Under
conditions of induction, however,
this speci-
ficity must
be
relieved.
Suppose, for example,
thatthere were 1000 molecules
of
repressorper
cell: only
0.04oh
of operators would be free
under conditions of repression. Upon induc-
tion, though, only 40o/o
of operators would
become
free. We therefore
see an inverse cor-
relation between the ability to achieve com-
plete
repression
and the ability to relieve
repression effectively. We assume that the num-
ber of
repressors
synthesized in
vivo
has been
subject to selective
forces
that balance these
demands.
The
difference
in
expression of the lactose
operon
between its induced and repressed
states
in wvo isactually
I
03x.
In
other
words,
even
when
inducer is absent, there is a basal level of expres-
sion of
-O.l%
of the induced level. This
would
be reduced if there were more repressor
protein
present
and
increased if there
were
less. Thus it
could be impossible to establish tight repression
if
there were
fewer repressors
than the ten
found
per
cell, and
it might
become difficult to
induce
the operon if there were too many. It is
possible
to
introduce t};re lac operator-repressor system
into the mouse. When Lhe lac operator is con-
nected to a tyrosinase
reporter
gene,
the enzyme
is induced by the addition of IPTG. This means
that the
repressor is finding its
target
in a
genome
l0r times
larger
than that of
E
coli.Induction
occurs
at approximately the same concentration
of
IPTG
as
in bacteria. We do not. however. know
the concentration of
Lac repressor
and how effec-
tively
the target is induced.
In
order
to extrapolate invivo fromthe affin-
ity of a DNA-protein
interaction
in vitro, we
need
to
know the effective concentration of DNA ln
vivo. T];re
"effective
concentration" differs
frt-rm
the mass/volume because of several
factors. The
effective concentration
is increased, for exam-
ple,
by molecular crowding, which occurs
when
polyvalent
cations neutralize
-90%
of
the
charges on DNA, and
the
nucleic acid collapses
into
condensed
structures.
The
major force
that
decreases the effective
concentration
is the inac-
cessibility of
DNA that
results
from occlusion
or sequestration by
DNA-binding
proteins.
One way to
determine
the effective
con-
centration
is to compare
the
rate of a
reaction
in
vitro
and
in vivo thar depends
on DNA concen-
tration. This
has been done
using
intermolec-
ular recombination
between
two
DNA
molecules. To
provide
a
control, the
same
reac-
tion is followed as an
intramolecular
recombi-
nation, that
is, the two
recombining
sites
are
presented
on the
same
DNA
molecule.
We
assume
that concentration
is the same
in
vivo
and in vitro for t]ne
intramolecular
reaction,
and
therefore
any difference
in the ratio
of inter-
molecular/
intramolecular
recombination
rates
can
be attributed
to a change
in the effective
concentrationinvivo.
The
results of such
a com-
parison
suggest
that the
effective
concentration
of DNA is reduced
>10-fold invivo.
This could
affect
the rates
of reactions
that
depend on DNA concentration,
including
DNA
recombination
and
protein-DNA binding.
It
emphasizes
the
problem
encountered
by all
DNA-binding
proteins in finding
their
targets
with sufficient
speed
and
reinforces
the conclu-
sion that diffusion
is
not adequate
(see
Fig-
ure I L22).
Repression
Can 0ccur
at
Multiple
Loc'
o
A repressor will act
on a[[ loci
that
have a copy
of
its
target
operator
sequence,
The lac repressor acts
only
on the
operator
of
tine lacZYA cluster.
Some
repressors,
however,
control dispersed
structural
genes
by binding
at
more than one
operator.
An example
is lhe
trp
repressor, which
controls
three
unlinked
sets
of
genes:
.
An
operator
at the
cluster
of structural
genes
trpEDBC,
controls
coordinate
s1m-
thesis
of the
enzymes
that synthesize
tryptophan
from
chorismic
acid.
.
An operator
at another
locus controls
tine aroH
gene, which
codes
for one
of
the
three enzymes
thatcatalyze
the ini-
tial
reaction
in the
common
pathway
of
aromatic
amino
acid
biosynthesis.
o
The trpRregulator
gene
is repressed
by
its own
product,
tlne
trp
repressor'
Thus
12.19
Repression
Can
Occur
at
Multipte
Loci 319
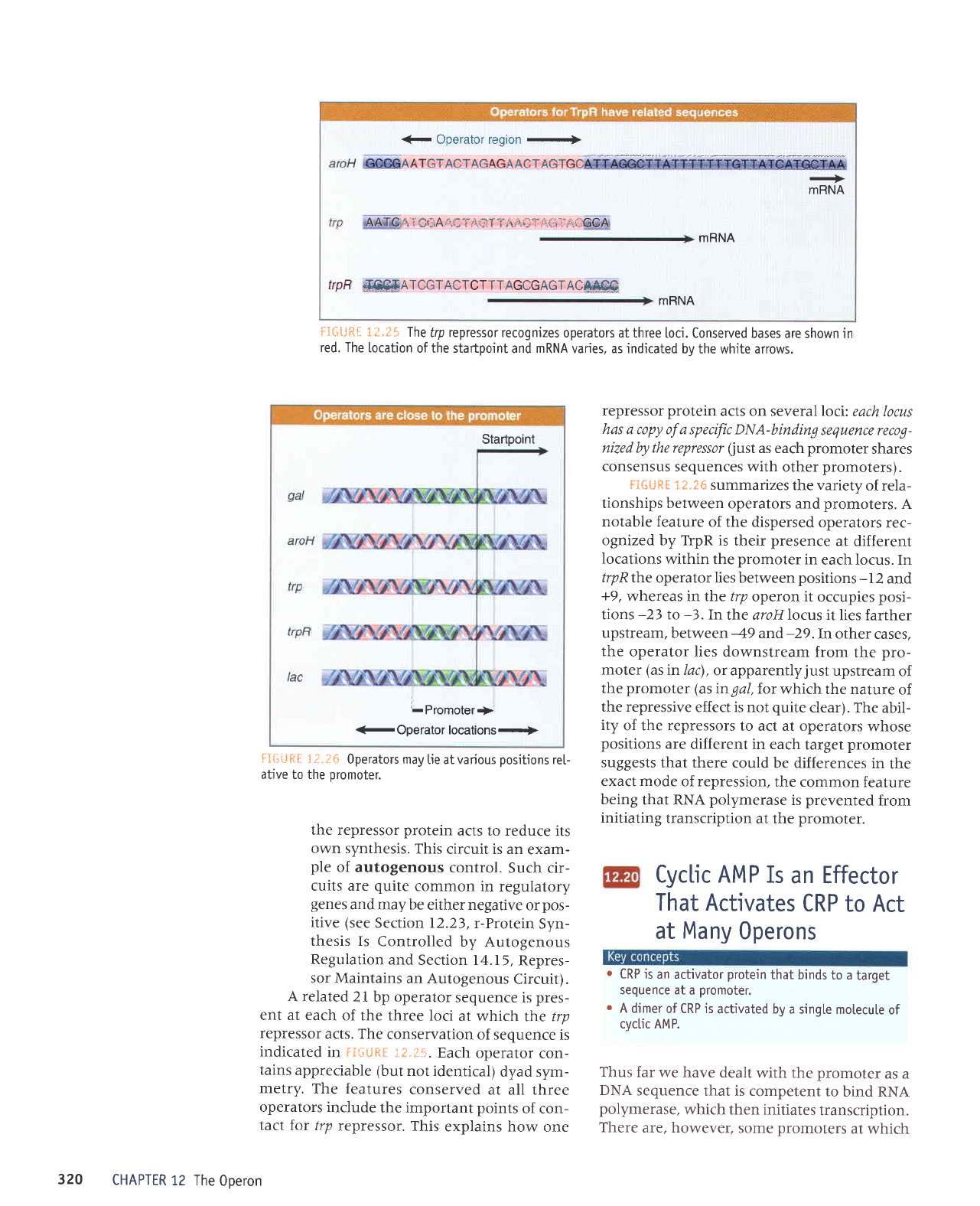
€ Operator region
+
+
mRNA
trp
AATCI'i''
.i:Arrr.:l
:1,i',
'
:
::
11,tl ]
.'r.i.:t'i
;!i,GCA
mRNA
frpF
ffiATCGTACTCTTTAGCGAGTAC*4@
mRNA
ri*iJftf
i?.t5 The
frp repressor recognizes
operators at three
loci. Conserved bases
are shown in
red. The
[ocation
of the startpoint
and
mRNA
varies, as indicated
bv the white arrows.
Startpoint
'-Promoter.i}'
3 Operat6y lq66lisns-r>
fiGiJtf 1i.I6
0perators may
lie at various
positions
rel
ative to the
promoter.
the repressor
protein
acts to reduce its
own synthesis.
This
circuit is an
exam-
ple
of autogenous
control.
Such cir-
cuits are
quite
common
in regulatory
genes
and may be
either negative
or
pos-
itive (see
Section 12.23,
r-Protein
Syn-
thesis Is
Controlled
by Autogenous
Regulation
and
Section l4.lr,
Repres-
sor Maintains
an Autogenous
Circuit).
A related
2i bp operator
sequence is
pres-
ent at each
of the
three loci
at which
the trp
repressor
acts. The
conservation
of sequence
is
indicated
in
FIiliJR[
:.?..J5.
Each
operator
con-
tains
appreciable (but
not identical)
dyad sym-
metry.
The
features
conserved
at all three
operators
include
the important
points
of con-
tact
for /rp
repressor.
This
explains how
one
CHAPTER
12 The
0oeron
repressor
protein
acts on several loci:
each locus
has a copy of a specific DNA-binding
sequence reclg-
nized by the repressor
(just
as each
promoter
shares
consensus
sequences with
other
promoters).
FtilLiRE
X?"?S
summarizes
the
variety
of rela-
tionships between
operators and
promoters.
A
notable feature
of the dispersed
operators
rec-
ognized by TrpR is
their
presence
at different
Iocations
within the
promoter
in
each locus. In
trpRlh.e
operator lies between
positions
-I2
and
+9,
whereas inthe trp
operon it
occupies
posi-
tions
-23
to
-3.In
the aroH locus it
lies farther
upstream,
between
-49
and-29.In
other
cases,
the
operator lies downstream
from
the
pro-
moter
(as
in lac)
,
or apparently
just
upstream
of
the
promoter (asingal,
for which
the nature
of
the repressive
effect is not
quite
clear).
The abil-
ity
of the repressors
to act at operators
whose
positions
are
different in each
target
promoter
suggests
that there
could be differences
in
the
exact mode
of repression,
the common
feature
being
that RNA
polymerase
is
prevented
from
initiating
transcription
at the
promoter.
Cyclic AMP
Is
an Effector
That Activates
CRP
to Act
at
Many
Operons
o
CRP is
an activator
protein
that binds
to a
target
sequence at
a
promoter.
r
A dimer
of CRP is
activated by a
single motecute
of
cyclic AMP.
Thus far
we have dealt
with the
promoter
as a
DNA
sequence
that is competent
to
bind RNA
polymerase,
which then initiates
transcription.
There
are, however,
some
promoters
at
which
320
