Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.

(see
Section
14.Il,
Repressor Uses a Helix-
T\rrn-Helix Motif to Bind DNA). Amino
acids
separated
by three
to
four
positions
lie on the
same face of an s-helix and are
therefore
in
a
position
to contact
adjacent base
pairs.
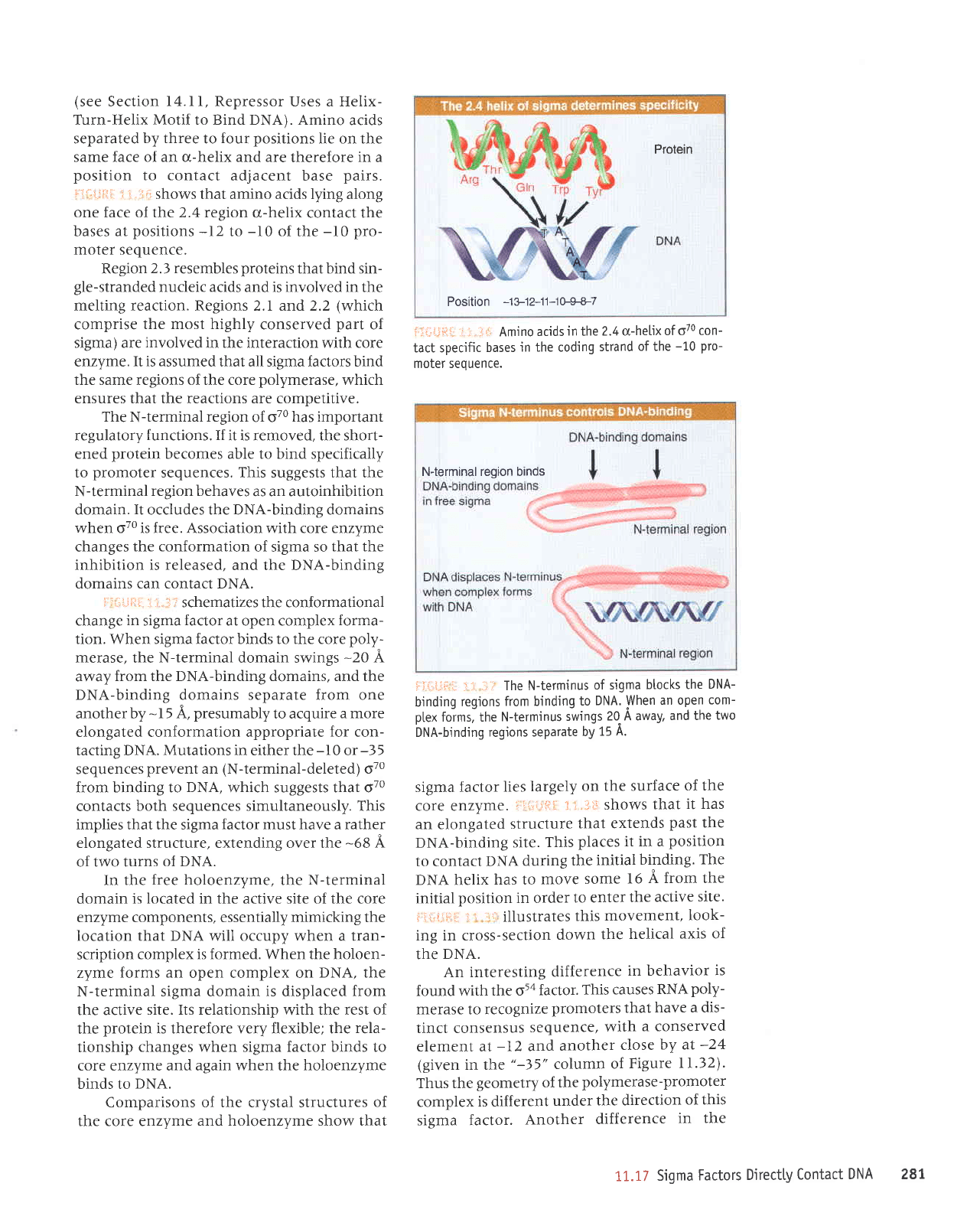
lril',l,jiil
5 i.:ri::
shows that amino acids lying along
one face of the 2.4 region
u-helix contact the
bases at
positions
-12
to
-10
of the
-10
pro-
moter sequence.
Region
2.3 resembles
proteins
that bind sin-
gle-stranded
nucleic acids
and
is
involved
in
the
melting reaction. Regions 2.1 and 2.2
(which
comprise
the most highly
conserved
part
of
sigma) are
involved
in the interaction with core
enzyme.
It is assumed
that all sigma factors bind
the same regions of the core
polymerase,
which
ensures that the reactions are competitive.
The
N-terminal
region
of o70 has
important
regulatory
functions. If
it is removed, the short-
ened
protein
becomes able to bind specifically
to
promoter
sequences. This suggests that the
N-terminal
region behaves
as an autoinhibition
domain.
It
occludes the DNA-binding domains
when o70 is free. Association with core enzyme
changes
the conformation
of sigma so
that the
inhibition
is released,
and the DNA-binding
domains can contact DNA.
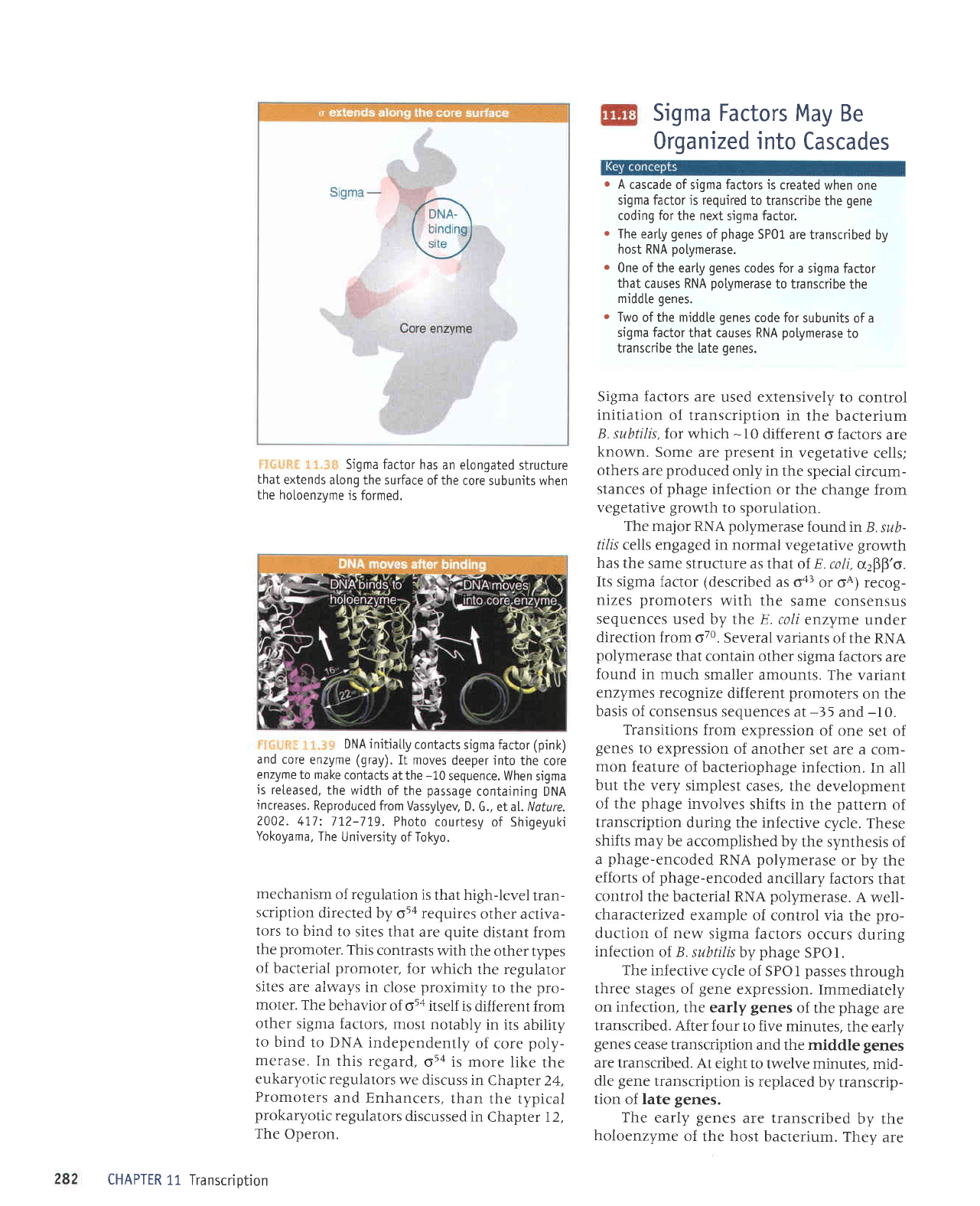
i:,i.-:-!iiil:
t::. ::
j-
schematizes the conformational
change
in
sigma
factor
at open complex
forma-
tion. When sigma factor binds to the core
poly--
merase, the N-terminal domain swings
-2O
A
away
from the DNA-binding domains, and the
DNA-binding domains separate from one
another by
-15
A,
presumably
to acquire
a more
elongated conformation appropriate
for
con-
tacting DNA. Mutations in either the
-
I
0 or
-3
5
sequences
prevent
an
(N-terminal-deleted)
o70
from binding to
DNA,
which suggests that o70
contacts
both sequences simultaneously. This
implies that the sigma
factor must
have a
rather
elongated structure, extending over the
-68
A
of two turns
of DNA.
In the
free holoenzyme,
the N-terminal
domain
is located in the active
site
of the core
enzyme components, essentially mimicking the
location that DNA will occupy
when
a tran-
scription complex
is formed. When the holoen-
zyme
forms an
open complex on
DNA, the
N-terminal
sigma domain is displaced
from
the active
site. Its relationship
with
the rest of
the
protein
is therefore very flexible; the
rela-
tionship
changes when sigma factor binds to
core enzyme and again when the
holoenzyme
binds to
DNA.
Comparisons
of the crystal structures
of
the core enzyme
and holoenzyme show that
Protein
Position
-1T12-11-1H-a-7
ilii-iililir
:l i,lr:r: Amino
acids
in
the
2.4 cr-helix
of o70 con-
tact specjfic
bases
in the coding
strand
of the
-10
pro-
moter sequence.
i:iir,i"iiii-
1.l
"-i,:
The N-terminus
of sigma
blocks the
DNA-
binding
regions
from binding
to
DNA. When
an open
com-
ptex
forms, the
N-terminus
swings
20. A away,
and the
two
DNA-binding
regions separate
by
15
A.
sigma factor
lies
largely
on the
surface
of the
core enzyme.
ir
i:i,iia;i:a
i i.;itr
shows
that
it has
an elongated
structure
that
extends
past
the
DNA-binding
site.
This
places it in a
position
to contact
DNA
during
the
initial
binding.
The
DNA helix
has to
move
some
l6
A from the
initial
position in order
to enter
the
active
site.
iii.i.ri:.i:
::
r'i
illustrates
this
movement,
look-
ing in cross-section
down
the
helical
axis of
the
DNA.
An interesting
difference
in behavior
is
found with
the o5a
factor.
This
causes
RNA
poly-
merase to recognize
promoters that
have a dis-
tinct consensus
sequence,
with
a conserved
element at
-12
and
another
close
by at
-24
(given
in the
"-35"
column
of
Figure
11.32).
Thus the
geometry of the
polymerase-promoter
complex
is different
under
the direction
of this
sigma factor.
Another
diff
erence
in the
Nterminal region binds
11.17
Sigma
Factors
Directly
Contact
DNA
287
Sigma factor has
an elongated
structure
that
extends
atong the surface
of the
core subunits when
the hoLoenzvme
is formed.
,
DNA initiatty
contacts sigma factor (pink)
and core
enzyme
(gray).
It moves
deeper into
the core
enzyme
to make
contacts
at the
-10
sequence.
When sigma
is
released,
the width
of
the
passage
containing DNA
increases.
Reproduced
from Vassylye4
D.
G., et al. Nature.
2002.
41.7: 71,2-71.9.
Photo
courtesy
of Shigeyuki
Yokoyama,
The
University
of
Tokyo.
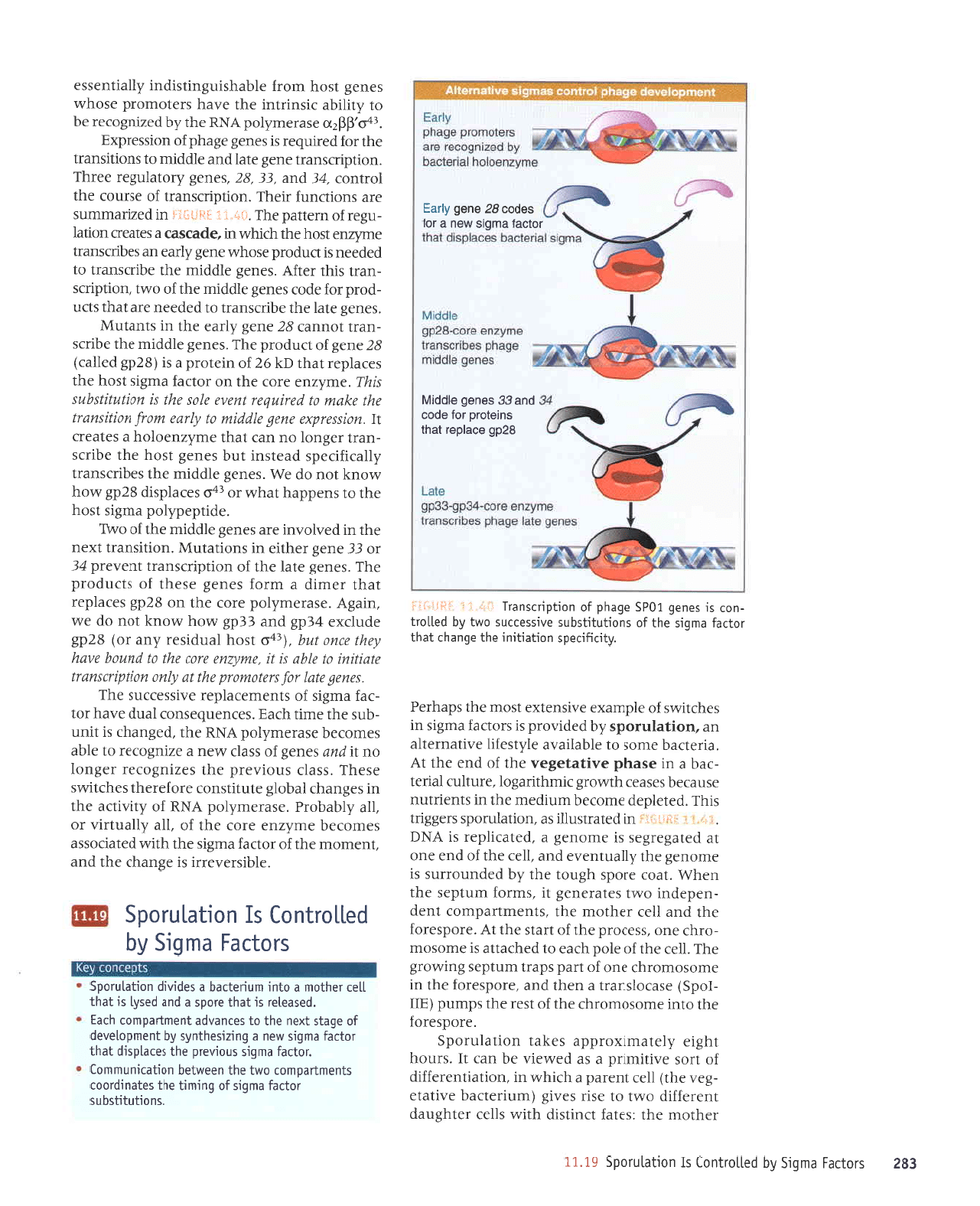
mechanism
of regulation
is
that high-level
tran-
scription
directed
by
otn requires
other
activa-
tors
to bind
to sites
that are
quite
distant from
the
promoter.
This
contrasts
with
the other
types
of
bacterial
promoter,
for which
the regulator
sites
are always
in
close
proximity
to
the
pro-
moter.
The
behavior
of o5a itself
is different
from
other
sigma
factors,
most notably
in its
ability
to bind
to DNA
independently
of
core
poly-
merase.
In
this regard,
o5a is more
like
the
eukaryotic
regulators
we discuss
in
Chapter 24,
Promoters
and
Enhancers,
than the
typical
prokaryotic
regulators
discussed
in
Chapter I 2,
Ihe
Operon.
CHAPTER
L1 Transcriotion
Sigma Factors
May Be
0rganized into
Cascades
.
A cascade of
sigma
factors is
created
when one
sigma factor is required
to transcribe
the
gene
coding for
the
next
sigma
factor.
.
The earty
genes
of
phage
SP01
are transcribed
by
host RNA
polymerase.
.
0ne of the earty
genes
codes for
a sigma factor
that
causes RNA
potymerase
to transcribe
the
midd[e
genes.
r
Two
of the midd[e
genes
code for
subunits of a
sigma
factor
that
causes RNA
polymerase
to
transcribe the [ate
genes.
Sigma factors are
used extensively
to
control
initiation
of transcription
in the
bacterium
B.
subtilis, for which
-10
different
o factors
are
known.
Some
are
present
in
vegetative
cells;
others are
produced
only in
the special
circum-
stances
of
phage
infection
or the
change from
vegetative growth
to sporulation.
The major RNA
polymerase
found in
B. sub-
tilis cells engaged
in normal
vegetative
growth
has the
same structure as
that of. E.
coli, szFF'o.
Its
sigma factor
(described
as oa3
or oA) recog-
nizes
promoters
with the
same
consensus
sequences
used
by the E. coli
enzyme
under
direction from
o70. Several
variants
of the RNA
polymerase
that contain
other sigma factors
are
found in
much smaller
amounts. The
variant
enzymes
recognize
different
promoters
on the
basis of consensus
sequences
at-35
and
-I0.
Transitions
from
expression
of one
set of
genes
to
expression of another
set
are a com-
mon feature
of bacteriophage
infection.
In
all
but the
very simplest
cases, the
development
of the
phage
involves
shifts in
the
pattern
of
transcription
during the infective
cycle. These
shifts
may be accomplished
by the synthesis
of
a
phage-encoded
RNA
polymerase
or by
the
efforts of
phage-encoded
ancillary
factors
that
control
the bacterial RNA
polymerase.
A
well-
characterized
example of
control
via
the
pro-
duction of new
sigma factors
occurs
during
infection
of. B.
subtilis by
phage
SPOL
The infective
cycle
of SPOl
passes
through
three
stages
of
gene
expression.
Immediately
on
infection,
the early
genes
of
the
phage
are
transcribed.
After four
to five minutes,
the early
genes
cease
transcription
and the middle genes
are transcribed.
At
eight to
twelve minutes,
mid-
dle
gene
transcription
is
replaced
by transcrip-
tion
of late
genes.
The
early
genes
are transcribed
by the
holoenzyme
of
the host bacterium.
They
are
E8Z
s.rollel
eu6rs nq pellolluol
sl
uoqelntods
6I.Il
JJqlou
eql
:sJleJ
t)urlsrp
qllM
sllal JalqSnpp
luJJaJJIp
oaa.l
ol esIJ seAIS
(runtratreq
JAIlple
-8a,t
aqt)
11ar
luared
e
qJrqM
ur
'uorlprlueJJJJrp
Jo
1:OS
anrlllUrrd
e sp
peMJrA
Jq upJ
1I
'slnoq
tq8ra
LlaterurxorddB
se>lpt uortelnrod5
'aJoosJloJ
Jqt olur JruosoluorqJ
eqt
Jo
lsal
Jr{t sdurnd
(91
-IodS)
JspJolsupJl
e uaqt
pue
'arodsaroJ
eql ur
JurosouroJqJ
auo
Jo
ged
sderl unldas
Sur.tror8
aqJ
'llal
aqt
yo
alod
q)ea
ot
pJqJplle
sr eurosour
-orqJ
euo
'ssarord
eql
Jo
upts
eqt
tv'arodsaro;
Jq1
pue
IIJJ
JJqtoru
Jqt
'sluJruuedruot
luap
-uadapur
o,lrl sJleraua8
1r
'sruro;
runldas aql
uJqM
'leor
arods
q8nol
aql ,{q
pepunorJns
sr
eruoue8 aql ,(llenruala pue
'lle)
Jql
Jo
pua
auo
te
pate8a,r8as
sr aruoua8 e
'paterrldar
sl
VNC
':
::
'
t
tri::::
j
UI
pelPJlsnll
Se
'uOIlelnJOdS
Sra38t.tl
srql
'palaldap
Jruo)Jq
unrpJru
aql ur stuJutnu
esneJJq
seseJJ
qlMoJB
rnuqlrreSol'JrnllnJ
IprJJl
-rpq
e ur
aseqd a,rllela8aa
aql
Jo
puJ
eql
tV
'erJelJeq
Jruos ot JlqelreAp
a1,{tsa;r1
JnupurJ}le
ue'uollelnrods.{q papraord
sr sJol)eJ
Bru8rs ur
sJrIJlrMS yo
aldruexa
anrsualxJ
tsoru
aql sdeqra4
'flLrrlLrads
uorlprlrut
eq1 a6ueqr
1eq1
ro1:e1
euErs
aql
Jo
suorlnlqsqns
alrssaltns
orvrl [q
pa11or1
-uor
st saua6
1665
abeqd
1o
uorldursueJl
r-:r:,'i
L
j;jil1tj-r:
'suorlnlqsqns
tolre; eu6rs
;o
6uLu4 aql saleurptooJ
SluAulUP0 tllol oMl eql
UaA/v\}aq uotleltu
n u U 0l
'rope1
euErs snonard
aq1 sareldsLp
1eq1
ro1re1
eurbrs
rvrau
e burzrsaqlur\s fiq
luaLudolanap
;o
ebels
lxou
aql ol soluplpp
luauleduor
qre3
'paspalaj
sr
1eq1
erods
e
pue
pesAl
sr
1eq1
ller
reqlour
e 01u! unuallpq
e
sopr^rp
uoqeln.lods
srollel
eu6r5 Aq
pollolluol
sJ uorlPlnrods
'JIqTsJJAJJJT
sr a8ueqr
eq1
pue
'lueruoru
Jql
]o
JolJpJ eru8rs
aqt
qllM paterlosse
sJruoreq aru.dzua
arol Jql
Jo
'll€
dllenpt.
ro
'11e
,{1qeqor4
'aseraruz(1od
VNU
Jo
{tr,rrDe
aql
ur sa8ueqr
1eqo13
Jlntrtsuo)
eroJJJJql
seqJlrMS
JSJqI
'sselJ
snorlard
aql sazruSolar
ra8uol
ou
tr
puv
saua8
;o
ssplJ Meu
e azruSorar
ol JIqe
seruoJaq
aseraru,(1od
vNu
Jql
'paSueqr
sl
llun
-qns
eql Jruu
qreg
'saruanbasuoJ
Ipnp
eleq Jol
-re;
eru8rs
Jo
slueruJteldar
a,Lrssartns
egJ
'saua6
a1a1 nt aaloutotd
aLll
p
[lul
uotldtttsuutl
awutul
q
ang s1
4
'autfzua
aJil a4t
q
punlq
aroLl
[a41atuo
fl4'Gvs lsoq
Ienprser
Iue,ro)
gTdB
epnlJXJ
7gd6
pue
EEdB
,r,roq Mou>l
tou
op e,lr
'ure8y
'aseraru,{.1od
Jro) eq1 uo
gTdB
sateldar
tpql
Jerurp e
ruJo] saua8 asaql
Jo
slJnpoJd
aq1
'saua8
alel aql
Jo
uorldrrlsuen
tuatatd
,{
ro
gg'aua8
raqlra ur suorlelnw
'uorlrsupJl
lxJu
Jqt ur
pJ^lo^ur
:re saue3
Jlpppu
Jrll
Jo
o^41
'apndad,{.1od
eru8rs
lsoq
aqt o1 suaddeq
lpq^l'
ro
,uo
sareldsrp
g7d3,noq
,r,rou>I
tou
op J714
'saua8
a1pp1ur
eql seqrJJSuBJl
z(11err;nads peJtsur
1nq
saua8
lsor{
Jqt
eqrrJs
-uerl
ra8uol
ou upJ
leq1
aurkuaoloq
e saleaJJ
tI'uotssatdxa
auaS
a1ppuu o1
Qtva
wotl uotysun4
aLfl a4vrlt o1
pattnbat
lua^a
alzs aLfi
st ulqruUsqns
sn41'atu[zua
eJoJ
Jql uo Jol]eJ
eru8rs
lsoq
aql
sareldar
lpql
q1I97;o
uralord
e sr
(g7dB
palpr)
97
aua8p
pnpord
aq1
'saua8
a1pp1u rql
eqrrrs
-ueJt
louueJ
97
aua8,1rea
aql
q
sluplnw
'saua8
alq
aql eqrJJsuerl
o1
pspeau
3Je
leql
stJn
-pord
ro; apor
saua8 epp1u
Jqt
Jo
omt
'uorldrJJs
-uprl
srqt ra1;y
'saua8
JIppIu rql
eqrrJsupJl
ol
pepJeu
sI
lJnpo;d
asoq,vr
aua8 dlrea
ue saqlrJsuerl
aru,{zua
roq
Jqlq)r-q,l.t
ur
€ppJspJ
p
salpeJJ
uorlel
-n8ar;o
uralled
aqJ
'+ir'1.i.
]ij{i**l.t
ur
pazrJeruurns
JJp suolDunJ
JIaqJ
'uorldrrtsuerl
Jo
JSJnoJ
Jql
Iorluor
'rc
pue'$'Se
'saua8
Lroleln8ar
aarql
'uorldrrlsuerl
aua8
a1e1
pup
Jlpptru
ol
suorllsupJl
eql JoJ
paJrnbar
sr saua8
aSeqd
yo
uorssardxg
'tf
ddzn
aseraruLlod
yNU
rql ,{q
pezruSora;
aq
ol ^lr1rqe
JrsurJlur
aql eneq sraloruord
esoqal
sauaB
lsoq
uroJJ Jlqeqsrn8urtsrpur,{lerluassa
g7d6
ecelder
1eq1
suregord iol epoc
pue69
seueO
elppryl
lolce; eur6rs itlou
e lol
sapoc
gZ
eueb [pe3
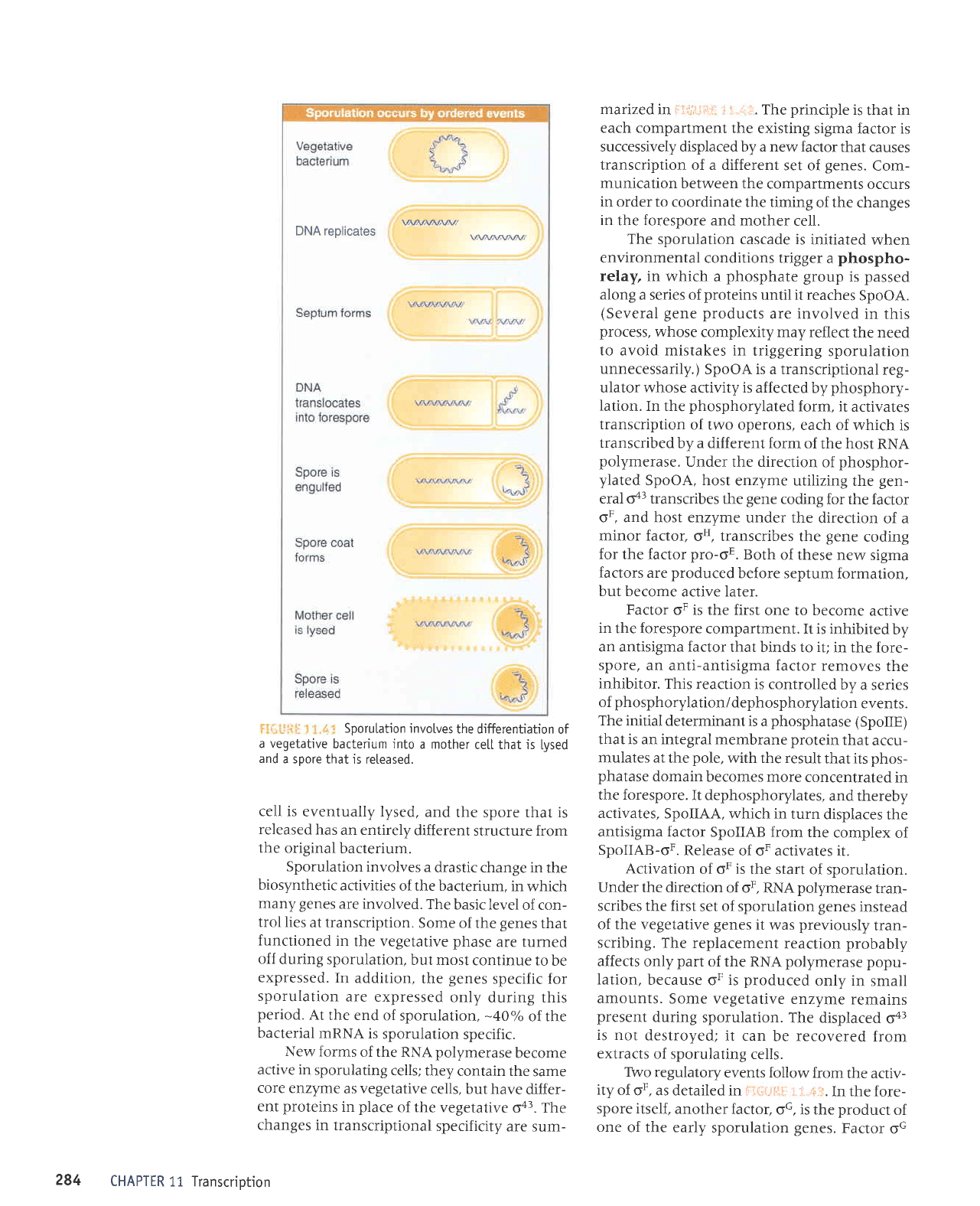
UAJ^/AI,\A]M/
DNA replicates
w
r:
,,':
.
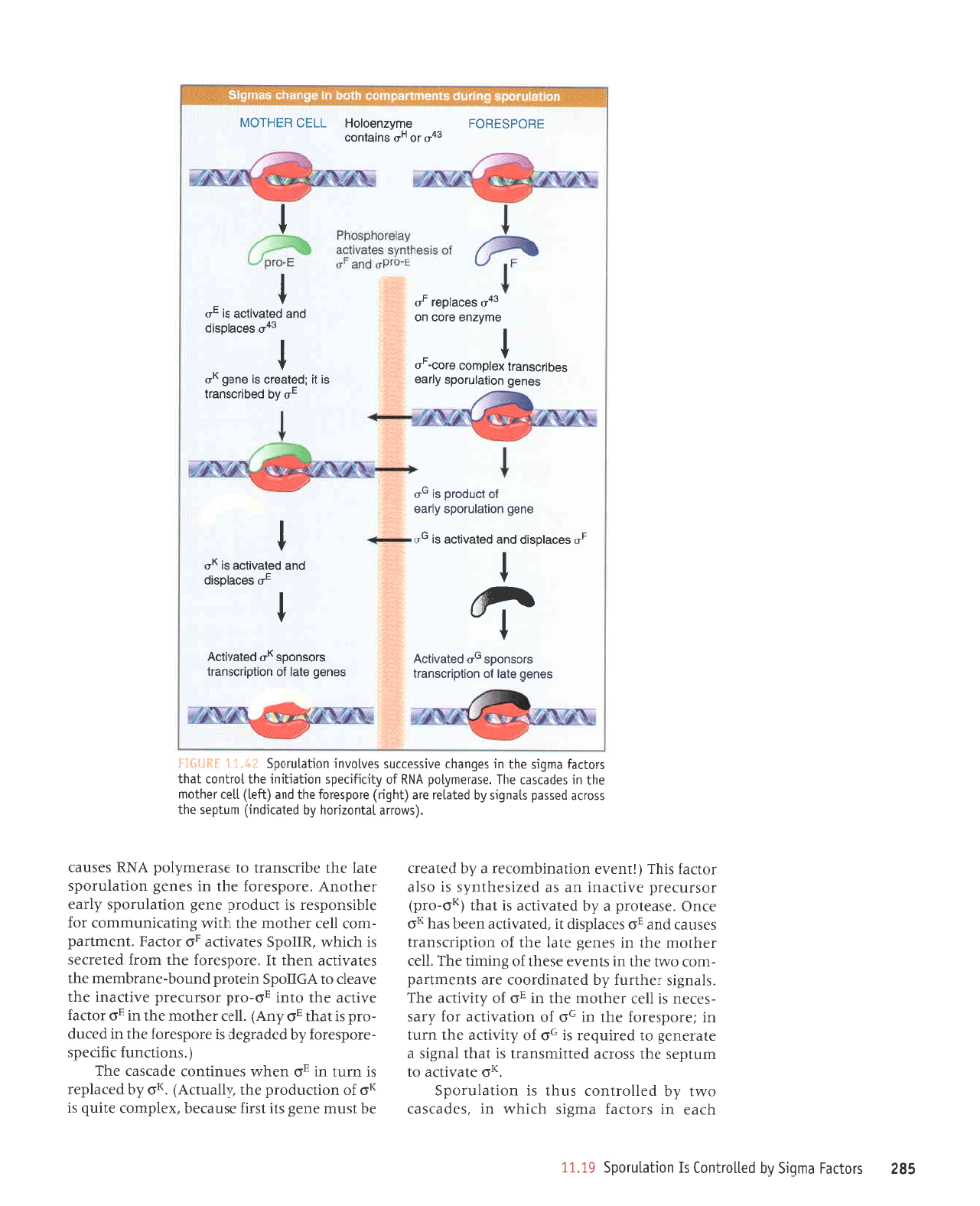
. Sporutation invotves
the differentiation
of
a
vegetative
bacterium into
a mother
cetl that is
tysed
and
a sDore
that is reteased.
cell is
eventually lysed,
and
the spore that is
released
has
an entirely
different
structure from
the original
bacterium.
Sporulation
involves
a
drastic change in
the
biosynthetic
activities
of the bacterium,
in
which
many
genes
are involved.
The
basic level
of con-
trol lies
at transcription.
Some of
the
genes
that
functioned
in
the
vegetative
phase
are turned
off
during
sporulation,
but most
continue to
be
expressed.
In
addition,
the
genes
specific for
sporulation
are expressed
only
during this
period.
At
the end
of sporulation,
-40%
of the
bacterial
mRNA
is
sporulation
specific.
New forms
of the RNA polymerase
become
::l'ff
;trJ:ll&Hlf
J:""',J?1',1*"Jft::
ent
proteins
in
place
of the
vegetative
o4l. The
changes
in transcriptional
specificity
are sum-
CHAPTER
1.1 Transcription
marized
in ijrli,ljiii I
j.r,.l'.
The
principle
is
that
in
each compartment
the existing
sigma factor is
successively displaced by a new factor
that
causes
transcription of a different
set of
genes.
Com-
munication between the
compartments
occurs
in order to
coordinate the timing
of the changes
in
the forespore and mother
cell.
The
sporulation cascade is
initiated
when
environmental
conditions trigger
a
phospho-
relay, in which
a
phosphate
group
is
passed
along
a series of
proteins
until it
reaches
SpoOA.
(Several gene
products
are involved
in this
process,
whose
complexity may
reflect the need
to avoid mistakes in
triggering
sporulation
unnecessarily.)
SpoOA is a transcriptional
reg-
ulator whose activity is
affected by
phosphory-
lation. In the
phosphorylated
form. it
activates
transcription
of two operons,
each of which
is
transcribed by a different form
of
the host RNA
polymerase.
Under the direction
of
phosphor-
ylated
SpoOA, host
enzyme utilizing
the
gen-
eral dl transcribes
the
gene
coding
for the factor
oF, and host enzyme
under the
direction
of a
minor factor,
oH, transcribes
the
gene
coding
for
the factor
pro-oE.
Both
of these
new sigma
factors
are
produced
before septum
formation,
but become
active later.
Factor
oF is the first
one to become
active
in
the forespore
compartment. It is inhibited
by
an antisigma factor
that binds to it;
in the fore-
spore,
an anti-antisigma factor
removes
the
inhibitor.
This reaction
is controlled
by
a series
of
phosphorylation/dephosphorylation
evenrs.
The initial
determinant is
a
phosphatase
(SpotrE)
that is an integral membrane protein
that
accu-
mulates at
the
pole,
with the result
that its
phos-
phatase
domain
becomes more
concentrated
in
the forespore.
It dephosphorylates,
and thereby
activates,
SpoIIAA, which
in turn
displaces
the
antisigma factor
SpoIIAB from
the
complex
of
SpoIIAB-oF.
Release
of oF activates
it.
Activation
of oF
is
the start
of sporulation.
Under
the direction
of oF, RNA
polymerase
tran-
scribes the first
set of sporulation genes
instead
of the vegetative genes
it
was
previously
tran-
scribing. The
replacement
reaction probably
affects
only
part
of the
RNA
polymerase
popu-
Iation,
because
oF
is
produced
only in
small
amounts.
Some vegetative
enzyme
remains
present
during
sporulation.
The displaced
oar
is not
destroyed;
it can
be recovered
from
extracts
of sporulating
cells.
TWo
regulatory
events follow
from
the activ-
ity
of
oF, as detailed in
i i
i;l,iii
!.'i
:t
i!ti..
In the fore-
spore itself,
another
factor,
oG. is the
product
of
one
of the early
sporulation
genes.
Factor
oc
284
982
slo]lel eu6rs tiq
pellorluol
sI
uorlelnrods
6I.II
qJeJ
ur sJolJeJ eruSrs
r{JIr{1!\
ur
'sJpeJSeJ
o.!tl ^q
pelloJtuo)
snql sr uortelnJods
')ro
Jle^rlJe Ol
runtdJs
Jqt ssorJp
pailltusueJl
sr
teqt
Ieu8rs
e
JleJJueB
ot
peJrnbJr
sr
eo
Jo
.{lr,,rrlre
Jql uJnl
ur
jarodsaroJ
Jqt
ul
eo
Jo
uorlelllre
ro; z(res
-sJJau
sr
IIJJ
rJqloru aqt
ur
to
Jo
dlrnrue
aql
'sleu8rs
requnJ .{q
pateurprooJ
JJe sluaruued
-ruof,
o^tt
Jql ur sluJAJ
esJql
Jo
3u[url JqJ
'lle)
Jaqtou
eqt ur
sauaS a1e1 Jql
Jo
uorldrnsuerl
sJsnpf,
pue
Eo
sareldsrp
tl
'palenrDe
ueJq seq
>ro
J)uO
'JseJtord
e Aq
pJte^rlJe
sr
leqt
(yo-ord)
rosrnrard
enrlJeur up
sp
pazrseqlu^s
sr osle
rotlpJ
srqJ
(11ua,ra
uorleurquoJlar
e
z(q
pJleaJJ
eq
tsnu
aua8 slr
lsJrJ
esnpJJq
'xaldruor
elrnb sr
)o
Jo
uorlJnpord aqt [ilenrrv)
'xo
r(Q
pareldar
sr urnl ur
[o
uJq^t
sJnurluof, JpeJSe)
aqJ
('suotpunJ
lqoads
-arodsaroy
,{q
paper8ap
sr arodsaro;
Jqt ur
paJnp
-ord
sr
lpql
so
z(uy)
'1ar
JJqlou
eql ur
ao
JoDpJ
alrlJp aql olul
ao-ord
ros.rnrard
J^rlJpur Jqt
JAeJIJ
or
Vglods
uralord
punoq-JueJqrueu
eql
selenrlJe
ueqr
lI
'arodsaro;
eqt
rxoJJ
pJlaDJS
sI I{JIq,tr
'UIIodS
salelrtJe
{o
JolJed
'luaruued
-ruoJ
IIJJ
Jeqtou
aql
qq'lt.
Surlelrunruruor
ro;
alqrsuodsar sr
pnpord
aua8 uorlelnrods
.dFea
JJqtouV
'arodsaro;
eqt ur
saua8 uorlelnrods
JIPI Jql eqlrJsupJl
ot aseraruLlod
vNU
sJsnef,
'(smore
lpluozuotl
{q
palerLpur)
unldas eq1
ssone
passed
sleubrs fq
palela.l
ale
(1qEu)
arodseroJ aql
pup (1e1)
11ar
taqlou
eql
ur sapelspr eql
'asetaufi1od
y15
1o
Alouoads
uor]prlrut
aql
lo.lluot
lpql
stolrel
eLubrs aq1 ur
sebueqr olrssaltns sanlo^ut
uoqelntod5
It'i.l :*{:*ij
ln
w
t
lo
soceloslp
pue pole^lce
sr
0
seueb e1e;
1o
uorlducsuell
sJosuods
go
palp^rlcv
eue6 uorle;nrods {pee
lo
lcnporo
sr
0o
seueO
uorle;nlods
Ipee
saqucsueJl
xelduoc
oJoc-ro
A
I
I
eulzue
oJoc uo
eto
SocPldot
,o
seueb
ege;
1o
uo[ducsuerl
s.rosuods
ro
polenlcv
I
I
='saceloslP
pue pelP^rlce
sr
y3
t
I
=o
{q
peqltcsuerl
sr
lr
:polBeJc
sr eua6
ro
t
elo
socelos!P
pue polp^llce
s!
lo
I
t
el'lO
H!
Suleluoc
jHOdSjHOl
aulAzueololl
ltSC HlHlOt/!
BacteriaI
RNA
Polymerase
Terminates
at Discrete
Sites
r
Termination
may require both recognition
of the
terminator sequence in DNA
and the formation
of a
hairpin
structure in the RNA
product.
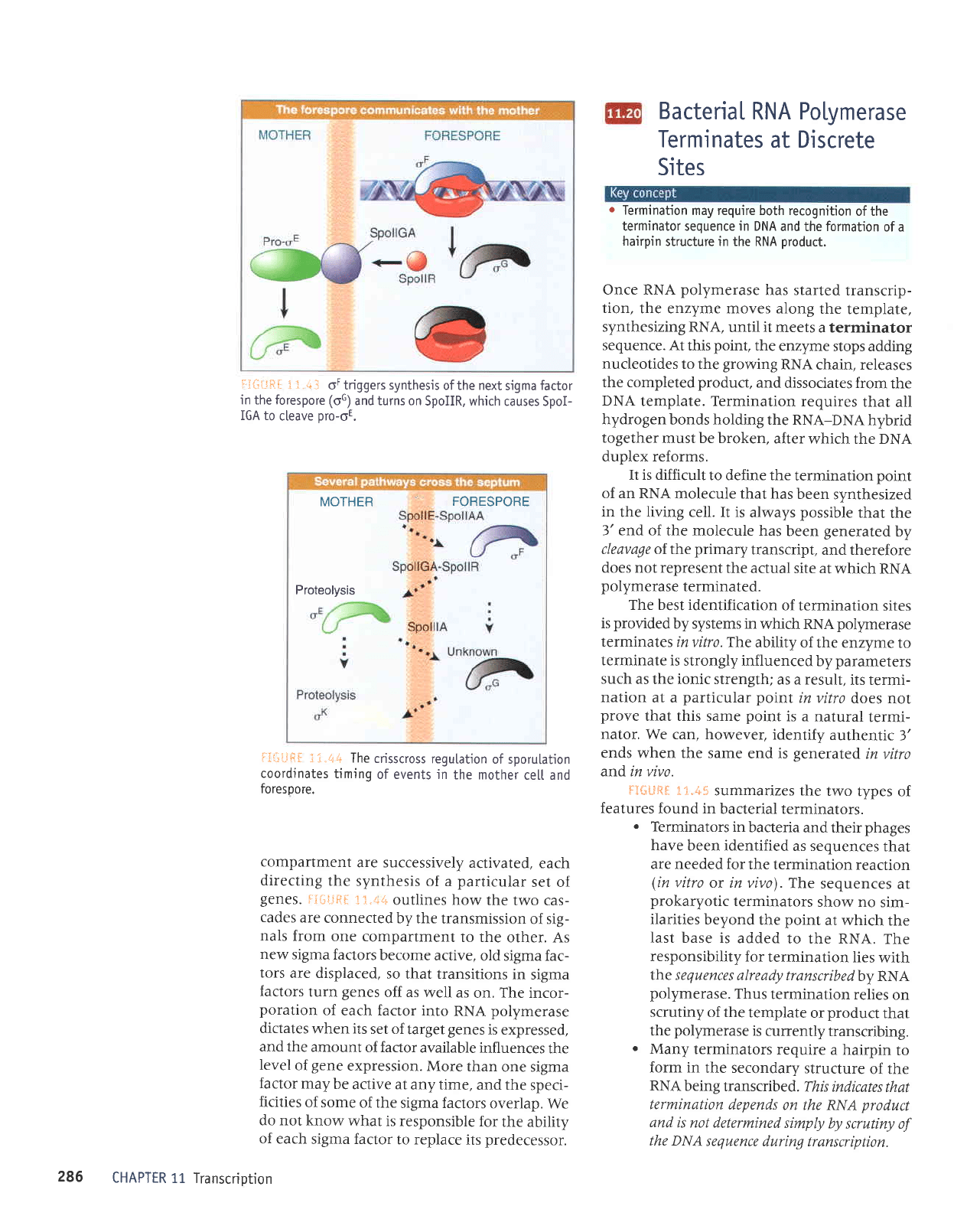
Once RNA
polymerase
has started
transcrip-
tion,
the enzyme moves along
the
template,
synthesizing RNA,
until it meets
a terminator
sequence.
At this
point,
the
enzyme
stops adding
nucleotides
to the
growing
RNA
chain, releases
the completed
product,
and
dissociates from
the
DNA
template. Termination
requires
that
all
hydrogen
bonds holding the RNA-DNA
hybrid
together
must be broken,
after
which the DNA
duplex reforms.
It is difficult
to define the
terminarion
point
of an RNA
molecule that has
been
synthesized
in
the living cell. It is
always
possible
rhat
the
3' end of
the molecule has
been
generated
by
cleavage of the
primary
transcript,
and therefore
does not represent
the actual
site at
which RNA
polymerase
terminated.
The best identification
of termination
sites
is
provided
by systems in
which RNA
poll.rnerase
terminates
invitro. The
ability
of the enzyme
to
terminate is
strongly influenced
by
parameters
such
as the ionic strength;
as a result,
its termi-
nation
at a
particular
point
in vitro
does
not
prove
that this same
point
is a
natural
termi-
nator.
We
can, however,
identify
authentic
J'
ends
when the same
end is
generated
in
vitro
and in vivo.
fi*LFR*
1.1-:i$
summarizes the
two
types of
features
found in
bacterial terminators.
.
Terminators
in
bacteria
and their
phages
have
been identified
as sequences
that
are needed for
the termination
reaction
(in
vitro
or in vivo).
The
sequences
ar
prokaryotic
terminators
show no
sim-
ilarities
beyond
the
point
at which
the
last base is
added
to the RNA.
The
responsibility
for
termination
lies
with
the sequences
already transcribedby
RNA
polymerase.
Thus
termination
relies
on
scrutiny of the
template
or
product
that
the
polymerase
is currently
transcribing.
.
Many
terminators
require
a
hairpin
to
form in
the secondary
structure
of the
RNA being
transcribed.
This indicates
that
termination
depends
on the RNA
product
and
is not determined
simply
by
scrutiny of
the DNA
sequence
during transcription.
ii*-*i
:
:.'t-:
oF triggers synthesis
of the next
sigma
factor
in the forespore (oG)
and turns on
SpoIIR, which
causes SpoI-
IGA to
cleave
oro-oE.
a-ILiii:s
::.ilr;
The
coordinates
timing
forespore.
crisscross
regutation
of sporulation
of events in
the mother
ce[[ and
compartment
are successively
activated,
each
directing
the synthesis
of a
particular
set of
genes.
-i!Liq{
::..++
outlines how
the
two cas-
cades
are
connected
by the
transmission
of sig-
nals
from
one compartment
to the
other. As
new sigma
factors
become
active,
old sigma fac-
tors
are displaced,
so that transitions
in
sigma
factors
turn
genes
off as
well as
on. The incor-
poration
of
each factor into
RNA
polymerase
dictates
when its
set of
target
genes
is expressed,
and the
amount
of factor
available
influences
the
level
of
gene
expression.
More
than
one sigma
factor
may be
active at
any time,
and the
speci-
ficities
of some
of the
sigma factors
overlap.
We
do not
know
what is responsible
for
the ability
of each
sigma factor
to replace
its
predecessor.
CHAPTER
11 Transcription
MOTHER
':
FORESPORE
Proteolysis
286
L8Z
Llos
'J
ur s.rolpululal
Jo
sadfl
oMf elv elaql
IZ'lI
'sanprsar
n
J0
unr
p
Aq
peMolloJ
sr
pue
uorbar
qru-l-g
p
sapnllur atnllnlls dool-Lua1s aql'dq
0Z
ol
/
ulolJ
q16ua1
u1 6ur[re,r surdrteq uloJ
leql
suorbar
lruro.lpurtp0
apnllur sloleurutal
lrsuulul
s?.I
t
It**3Ij
unr-p
pepuerls-a16urg
'&pssarau
eq
Aeu
ypg
aq1 uL
urdtreq
p
Jo
uorleulol
'aluanbas
lolpuru.lal
aq1;o ueatlsdn
palprol
aie uorJ
-pulu.lal
ro;
patrnbar
saruanbas
VN6
aq1
t?"t
I
3g{j$i*
aql
le
uofal
p
pue
'arnlJnJls
drepuolas
aq1 u1
utdrteq
p
:$*"tt
jHfr*Ij
ul
luaplne
sarnteeJ
Iernl
-JnJls
o^1,l eql J^Pq sJoleururral
Jrsurrlul
'0ltt
ut uorT
-eurruJJl
ur
pa^Io^ur
sr JolJpJ
aql
leql
Moqs
suorlptnur
pup
'zJlttut
(d)
rofrey
oqJ
Jo
uonrppe roJ
pJJU
Jql ^q
pJurJ
-ep
JJe sJoleurruJal
luapuadap-o{ll
.
'sJolPuFrrJal
)IsuIJlrrI
pJIIeJ
are sJlrs esJqJ
'JolJeJ
JJqIO ^ue
Jo
eJuesqP aql
q
salls urpuaJ
le
uilA r// JleurultJl uPf, er.u^zua Jro)
.
:0J1tA
Ut JleurruJal 01 SJOllpJ
leuorllppe
z(ue sarrnbar aseraur,{.1od
VNU
raqlJqM 01 8ur
-pJoJf,e
llu'g
ur
paqsrn8urlslp
JJe sJolpurruJeJ
's.rnlro
uorlPuruilal
qllq/{
ur uorber
qrp-n
p
r\q
pamolo;
pnpord
VNU
aql ul
uroJreq
qlu-l-9
P
J0
lsrsuol
sroleulurel lrsuuful
e
llD'l
srolPur.rulal
Jo
ut
peseolor
ere
VNH
pue
eseteuf;od
y1r1g
t
I
perrnber
eq [eu.r
'
vNU
ur urorBH
L---------T---------J
uorbe.r
poqucsuerl
ul
orP uorlEUrtlrjol
ro;
perrnbaj
secuanbos
llv
sadAl oMl arv eraql
'VN11
ul seJuenbes;o
uorlruSolar
aql se
IIJM
sB sJolJpJ .{relpue ,{q
ro asereru
-.{1od
y1qg,{q
tdrnsuert
aqt ul sleuErs
Jo
uorlru
-3o;ar
sellonur
luJle
uorleurruJal
eql
'q8noqr
'y51q
xaldnp
pup
aseraru,{1od
VNU
ueeulaq
uorlJprJlur
aqt uodn
,{1a1os
serlJr
uorlprlrur
seJrJqM'aseraur,{1od
er{l
Jo
surroJ
a,l,r1eura1p Lq
paqslldruorre
are.daql
'DpJ
uI'eu^zuJ JroJ
aql
qllM
Deretul
o1 sutalord
puonrppe
arrnbar
qloq
pue
'(uotleurruJJl
1p
uorlerJossrp
VN(-VNU
pue
uorlprtrur
te
VNO
Jo
3ur11aru
Iplllul)
spuoq uaEorpdq;o
Suqearq eJrnber
qtog
'uol1
-purruJat
pup
uorlplUur ur
padoldrxJ
sruJls^s
aql uaeMlaq s1a1p:ed Sunsaralur
aJe JJJqJ'loJl
-uor
rr;nads o1
palqns
eJe
qtoq (uorleurrural)
1l
ruorJ seler)ossrp
ro
(uor1e11pl)
vN(
r{t!r,r sare
-rJosse
aseraru,{1od
VNU
uJr{,lt. sa8els
aql snqJ
'uorssardxa
aua8
lorluor
o1 ,hrunuoddo
ue se
1nq
'JInJJIoru
VNU
eql
Jo
puJ
,€
aql Surleraua8
JoJ rusrupqJaru e
se
,{1drms
lou 1l
preBJJ
lsntu
JM
'tuJla
uorleurural aql,
Eurqreordde
u1
'(suopoJ
uollPuluJel
;o
uorssarddns s,aruosoqrJ
p
aqrJJsap
01
'Jpo)
aql
Jo
Supeag
adiJ-p111
qlpzr
aladruo) IeW
srossarddng
'7I'6
uorlJJS
ur
pesn
rurat
arups
aq1)
qEnoJqlppal pJIIeJ
lua^e
uB
'aruanbas
JoleulluJel
aq1
lsPd
uotldursuerl
anurluoJ ol
aur^zua Jqt sJSnpJ
uollpulruJallluv
'eseJaur
-,{1od
y}.Ig
qtl,ra
lJpJalul
leqr
sJolJpy ,{.re1prue
rr;rrads trqpaluarctd
aq upJ
lualJ
uorleurruJel
aq1
'sJoleurrural
aruos
1v
'uoJl€uruJJl
Jo
salJ
-uJrJrJJJ
rreql
ul
,{1apun
,,(ren
sroleurural

very
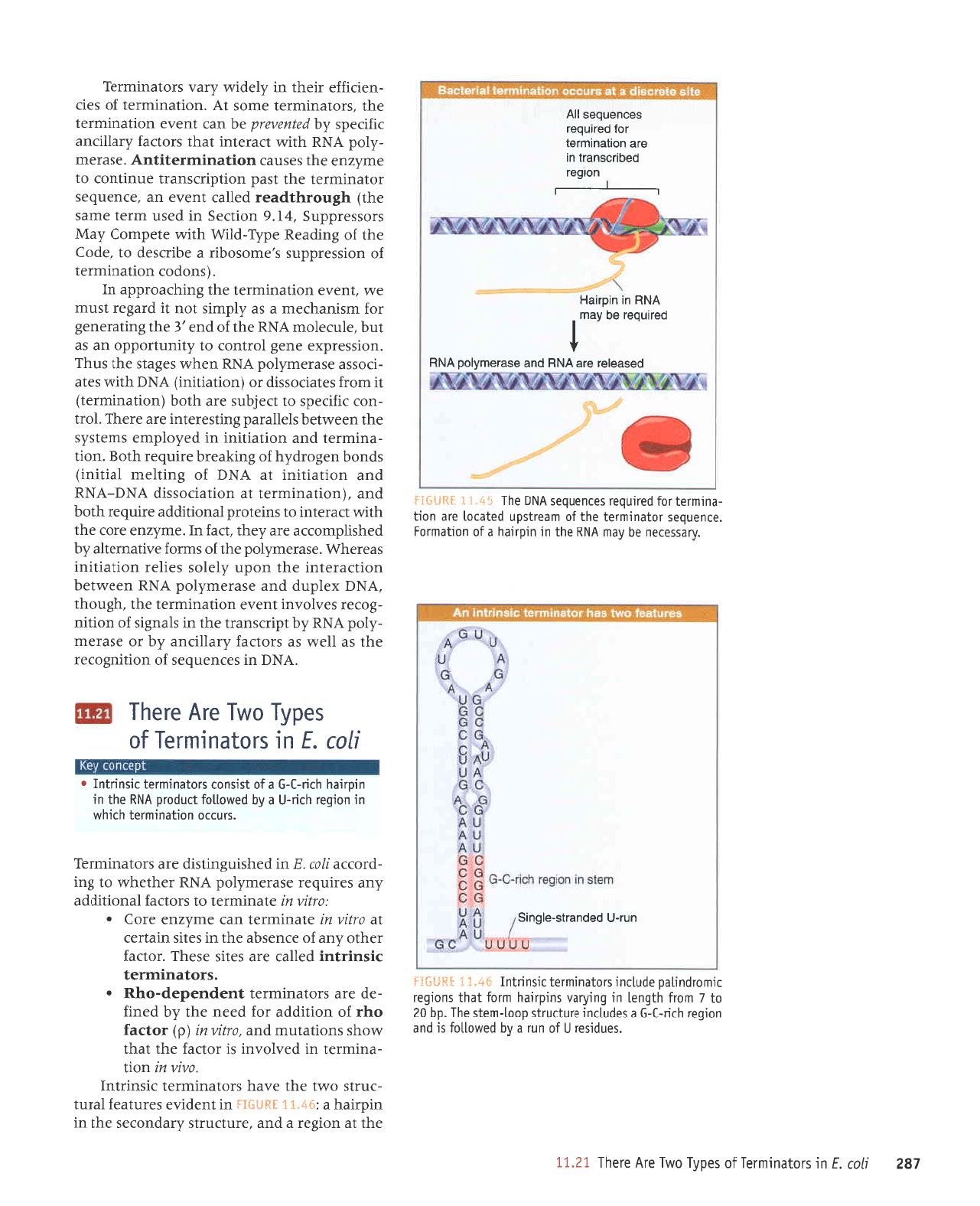
end of the
unit that is rich
in U residues.
Both features
are needed for
termination. The
hairpin
usually
contains
a G-C-rich region
near
the
base of the
stem. The
typical distance
between
the hairpin
and
the U-rich region
is
seven to nine
bases. There
are
-l
100
sequences
in
the E.
coli
genome
that fit these
criteria, which
suggests that
about half
of the
genes
have
intrin-
sic
terminators.
Point
mutations
that
prevent
termination
occur
within the
stem region
of the hairpin.
What is
the effect
of a hairpin
on transcription?
It is likely
that
all
hairpins
that
form in
the RNA
product
cause
the
polyme
rase
to
slow
(and per-
haps
to
pause)
in
RNA synthesis.
Pausing
creates
an opportunity
for termina-
tion to
occur. Pausing
occurs
at sites
that
resem-
ble
terminators
but have
an increased
separation
(typically
ten
to eleven
bases)
between the hair-
pin
and
the U-run.
If the
pause
site
does not
correspond
to a terminator,
though,
the enzyme
usually
moves
on again
to continue
transcrip-
tion.
The length
of the
pause
varies,
but
at a
typical
terminator
lasts
-60
seconds.
A downstream
U-rich region
destabilizes
the RNA-DNA
hybrid
when RNA
polymerase
pauses
at the hairpin.
The
rU-dA RNA-DNA
hybrid
has an
unusually
weak base-paired
struc-
ture;
it requires
the
least
energy
of any
RNA-DNA
hybrid to
break
the association
between
the two
strands.
When
the
polymerase
pauses,
the RNA-DNA
hybrid
unravels from
the
weakly
bonded
rU-dA
terminal region.
Often,
the actual
termination
event
takes
place
at any
one of
several
positions
toward
or at
the
end
of the U-rich
region,
as though
the enzyme
"stutters"
during
termination.
The
U-rich region
in
RNA
corresponds
to
an A-T-rich
region in
DNA,
so we
see that A-T-rich
regions
are impor-
tant
in intrinsic
termination
as well
as initiation.
Both
the sequence
of
the hairpin
and
the
length
of
the U-run
influence
the
efficiency
of
termination.
Termination
efficiency
in vitro,
however, varies
from 2o/o
lo 90oh
and
does not
correlate
in any
simple
way with
the constitu-
tion
of the hairpin
or
the number
of
U
residues
in
the
U-rich region.
The
hairpin
and
U-region
are therefore
necessary,
but not
sufficient,
and
additional
parameters
influence
the interaction
with
RNA
polymerase.
In
particular,
the
sequences
both
upstream
and
downstream
of
the intrinsic
terminator
influence
its
efficiency.
Less is
known
about
the signals
and
ancil-
lary
factors
involved
in termination
for eukary-
otic
polymerases.
Each
class
of
polymerase
uses
a different
mechanism
(see
Chapter
26,
RNA
Splicing
and Processing).
Transcri
ption
How
Does
Rho Factor
Work?
o
Rho factor is
a terminator
protein
that
binds to a
ruf
site on nascent
RNA and tracks
atong
the RNA
to
retease
it from
the RNA-DNA hvbrid
structure
ar
the RNA
potymerase.
Rho
factor is an
essential
protein
in E.
coli that
functions
solely
at the stage
of termination.
It
acts at rho-dependent
terminators,
which
account
for about half
of E. coli
Lerminators.
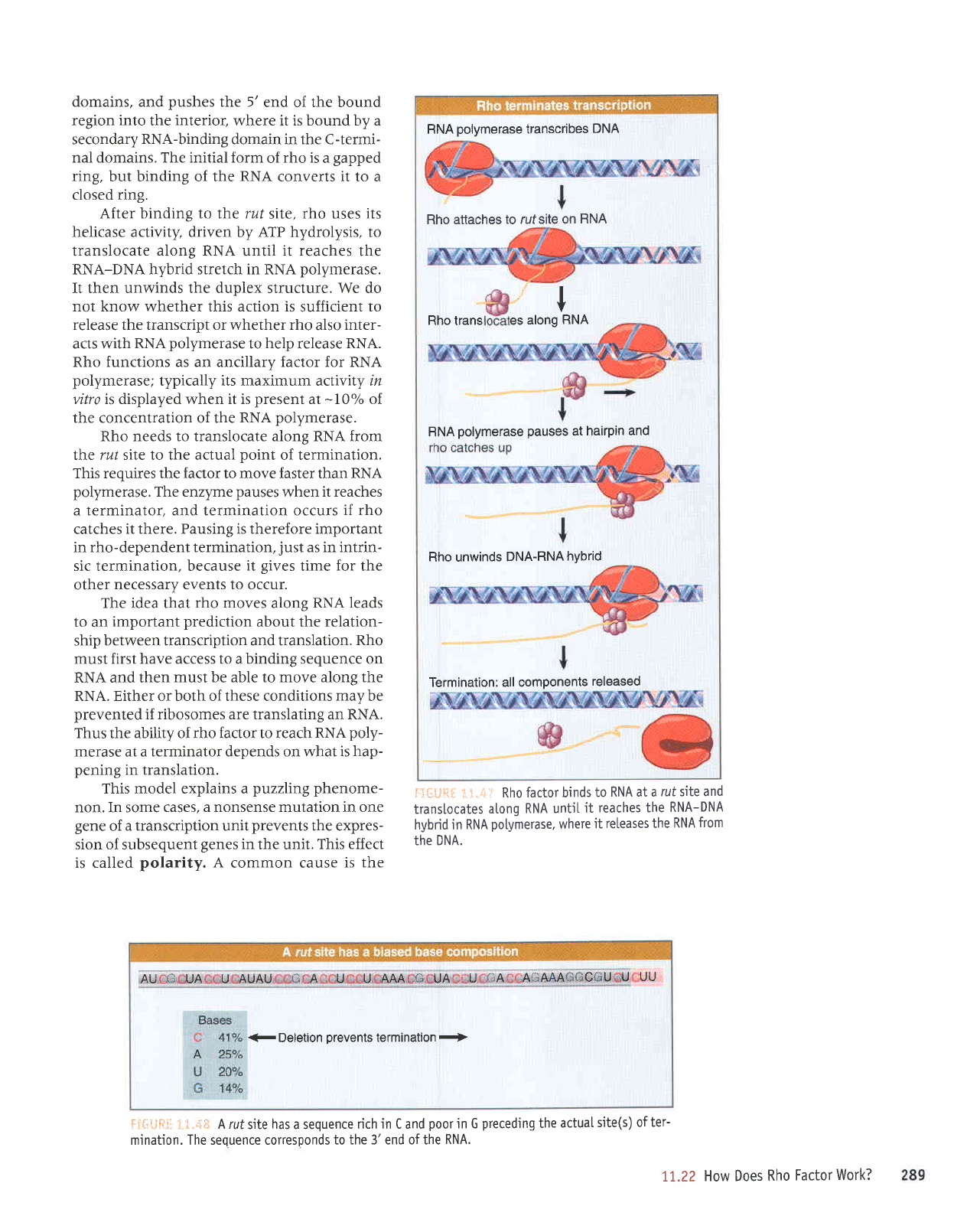
FIGURE x t
-4? shows how
rho functions.
First
it
binds to
a sequence within
the
transcript
upstream
of the
site of termination.
This
sequence
is called
a rut site
(an
acronym
for
rho
utilization).
The rho then
tracks
along
the
RNA
until it catches
up ro RNA
polymerase.
When the RNA
polymerase
reaches
the
termi-
nation
site, rho
acts on the RNA-DNA
hybrid
in
the enzyme
to cause release
of the
RNA. Paus-
ing
by the
polymerase
at
the site
of termination
allows
time for rho factor
to translocate
to the
hybrid
stretch
and is an important
feature
of
termination.
We
see
an
important general
principle
here.
When
we
know
the
site on DNA
at which
some
protein
exercises its
effect, we
cannot
assume
that
this coincides
with the DNA
sequence
rhat
it
initially recognizes.
They
can
be separate,
and
there
need not
be a fixed relationship
between
them. In f.act,
rut sites in
different
transcription
units are found
at varying
distances preceding
the sites
of termination.
A similar
distinction
is
made
by antitermination
factors (see
Sec-
tion 11.24, Antitermination
Requires
Sites That
Are Independent
of the Terminators).
The
common feature
of. rut
sites is
that
the
sequence is
rich in
C
residues
and
poor
in
G
residues
and has no
secondary
structure.
An
example
is
given
in FSGttRil
it.4S.
C is by far
the
most
common
base
(4lo/o)
and
G is
the least
common
base
(Ia%|.
rut
sites vary in
length.
As
a
general
rule,
the efficiency
of
a rutsile
increases
with
the length
of the
C-rich/G-poor
region.
Rho is a member
of
the family
of hexameric
ATP-dependent
helicases.
The
subunit
has
an
RNA-binding
domain
and an
ATP hydrolysis
domain. The
hexamer
functions
by
passing
nucleic
acid through
the
hole in
the middle
of
the
assembly
formed from
the RNA-binding
domains
of
the subunits.
f,cfruRg
11.4s
shows
that
the structure
of rho
gives
some indications
of how
it functions.
It
winds
RNA from
the
3'
end
around
the
exterior
of the
N-terminal
CHAPTER
11
domains, and
pushes
the 5'
end of the bound
region
into
the
interior,
where it is bound by a
secondary
RNA-binding
domain in the C-termi-
nal domains. The initial form
of
rho is
a
gapped
ring,
but binding of the
RNA
converts it to a
closed ring.
After binding to
the
rut
site, rho uses
its
helicase activity, driven by ATP hydrolysis, to
translocate
along RNA
until
it reaches
the
RNA-DNA hybrid stretch in RNA
polymerase.
It then unwinds the duplex structure. We do
not know whether this action is
sufficient
to
release the transcript or whether rho
also
inter-
acts with
RNApolymerase
to help release
RNA.
Rho functions as an ancillary factor for RNA
polymerase;
typically its maximum aclivity in
vitro is displayed when it is
present
at
-l0o/o
of
the concentration of the RNA
polymerase.
Rho needs to translocate along RNA from
thre rut site to the actual
point
of termination.
This requires the
factor
to move faster than RNA
polymerase.
The enzl.rne
pauses
when
it reaches
a terminator, and termination occurs if rho
catches
it there. Pausing is therefore important
in
rho-dependent termination,
just
as in intrin-
sic termination, because
it
gives
time
for
the
other
necessary events to occur.
The idea that
rho moves
along
RNA leads
to an
important
prediction
about
the
relation-
ship
between transcription and translation. Rho
must first have access to a binding sequence on
RNA and then
must be able to move along the
RNA. Either or both of these conditions
may
be
prevented
if ribosomes are translating an RNA.
Thus
the
ability of rho factor to reach
RNA
poly-
merase
at
a terminator depends on what is hap-
pening
in translation.
This model explains apluzzlir'g
phenome-
non. In some
cases, a nonsense mutation in one
gene
of
a transcription unit
prevents
the expres-
sion of subsequent
genes
in
the unit.
This effect
is called
polarity.
A
common cause
is the
iI+i.il:l i i.ri.j Rho
factor binds
to
RNA at a
ruf site and
trans[ocates
atong
RNA untjt
it
reaches the
RNA-DNA
hybrid in RNA
poLymerase,
where
it
reteases the
RNA
from
the
DNA.
AU * UA U
AUAU
*
A U U
AAA.*
UA.]
U
*A AfiAAA**C*U..U
UU
€ Deletion
prevents
termination +
l:$liFi:li.+S
ArufsitehasasequencerichinCandpoorinGprecedingtheactuaIsite(s)
ofter-
mination. The seouence corresoonds
to the 3'end of the
RNA.
RNA
polymerase
transcribes
DNA
Rho attaches
to ruf site on
RNA
I
V
Rho lrans
es
along BNA
RNA
polymerase
pauses
at
hairpin and
Rho unwinds
DNA-FINA
hybrid
Termination: all components
released
17.22
How Does
Rho
Factor
Work?
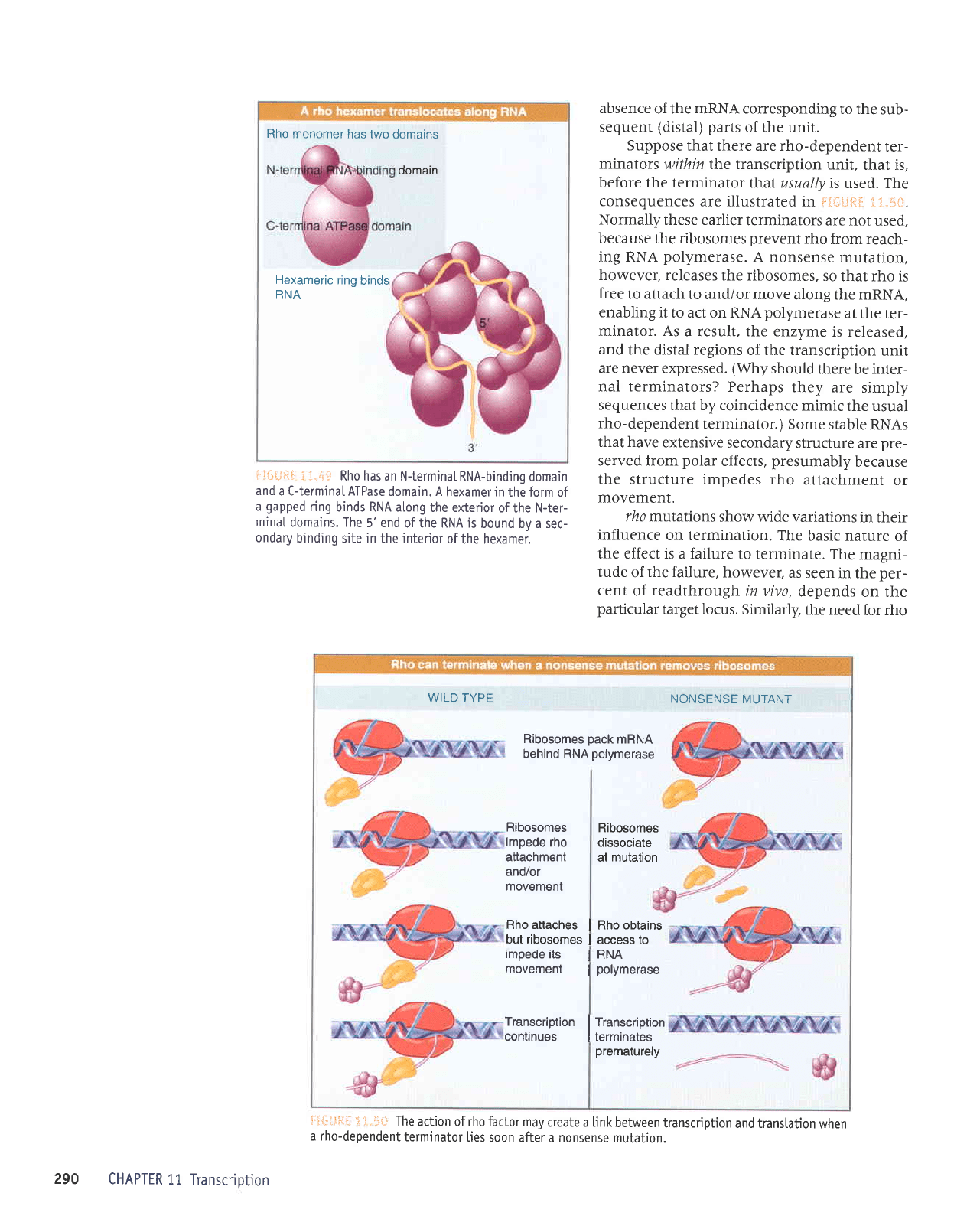
Rho monomer
has two
domains
domain
Hexameric
ring
binds
RNA
iiiL:il
: :.+"s
Rho has an N-terminaL
RNA-binding
domain
and
a C-terminaI ATPase
domain. A
hexamer in
the form of
a
gapped
ring
binds RNA along
the exterior
of the N-ter-
mjnal
domains. The
5'end
ofthe RNA is
bound by a sec-
ondary
binding
site in the
jnterior
of
the hexamer.
absence
of the nRNA corresponding
to the
sub-
sequent
(distal) pafis
of the unit.
Suppose
that there are rho-dependent
ter-
minators
within the transcription
unit,
that is,
before the terminator
that usually
is used. The
consequences
are illustrated
in
n:{*{t*
11,5*.
Normally these earlier
terminators
are not
used,
because the ribosomes
prevent
rho
from reach-
ing RNA
polymerase.
A nonsense
mutation,
however, releases
the ribosomes,
so
that rho is
free
to attach
to and/or move
along the mRNA,
enabling
it to act on RNA
polymerase
at the ter-
minator. As
a result, the enzyme
is released,
and the distal regions
of the transcription
unit
are never expressed.
(Why
should
there
be
inter-
nal terminators?
Perhaps
they are
simply
sequences
that by coincidence
mimic
the usual
rho-dependent
terminator.)
Some stable
RNAs
that have
extensive
secondary structure
are
pre-
served from
polar
effects,
presumably
because
the structure impedes
rho
attachment
or
movement.
rho mutations
show wide
variations
in their
influence
on termination.
The
basic nature
of
the effect is
a
failure
to terminate.
The
magni-
tude
of the
failure,
however,
as seen
in the
per-
cent
of
readthrough
in
vivo,
depends
on the
particular
target locus.
Similarly,
the need
for rho
WILD TYPE
NONSENSE
MUTANT
Ribosomes
pack
mRNA
behind
RNA
polymerase
Ribosomes
impede
rho
attachment
and/or
movemenl
Ribosomes
dissociate
at mutation
Rho attaches
I
Rho obtains
but ribosomes
I
access to
impede
its
I
RNA
movement
I
polymerase
Transcription
lTranscription
continues
I
terminates
prematurely
fl;#lJfti:'i'1.li#
Theactionofrhofactormaycreatealinkbetweentranscriptionandtranstationwhen
a rho-dependent
terminator
lies
soon after a nonsense
mutation.
290
CHAPTER
11 Transcription
