Knapp J.S., Cabrera W.L. Metabolomics: Metabolites, Metabonomics, and Analytical Technologies
Подождите немного. Документ загружается.

From Metabolic Profiling to Metabolomics 137
Another hybrid instrument used in the metabolic profile is the linear ion trap-triple
quadrupole mass spectrometer (Q-Trap) [157,158,168,212]. The Q-Trap mass spectrometer is
a modified triple quadrupole where the Q3 region can be operated either as a conventional
quadrupole mass filter or as a linear ion trap with axial ion ejection. Thus, the instrument
encompasses the functionality of an ion trap mass spectrometer, with its associated high
sensitivity for product ion scanning, and that of a triple-quadrupole mass spectrometer. The
system also has MS
3
capabilities which are useful to determine the origin of the fragments
[213]. The most powerful MS ion analyzers, in terms of resolution, are the FT-ICR
and the
Orbitrap; these instruments can also perform MS/MS. Nevertheless their application, coupled
with LC in metabolomics is rare [214-217], and an increasing number of applications,
especially for LC-Orbitrap can be possible in a near future.
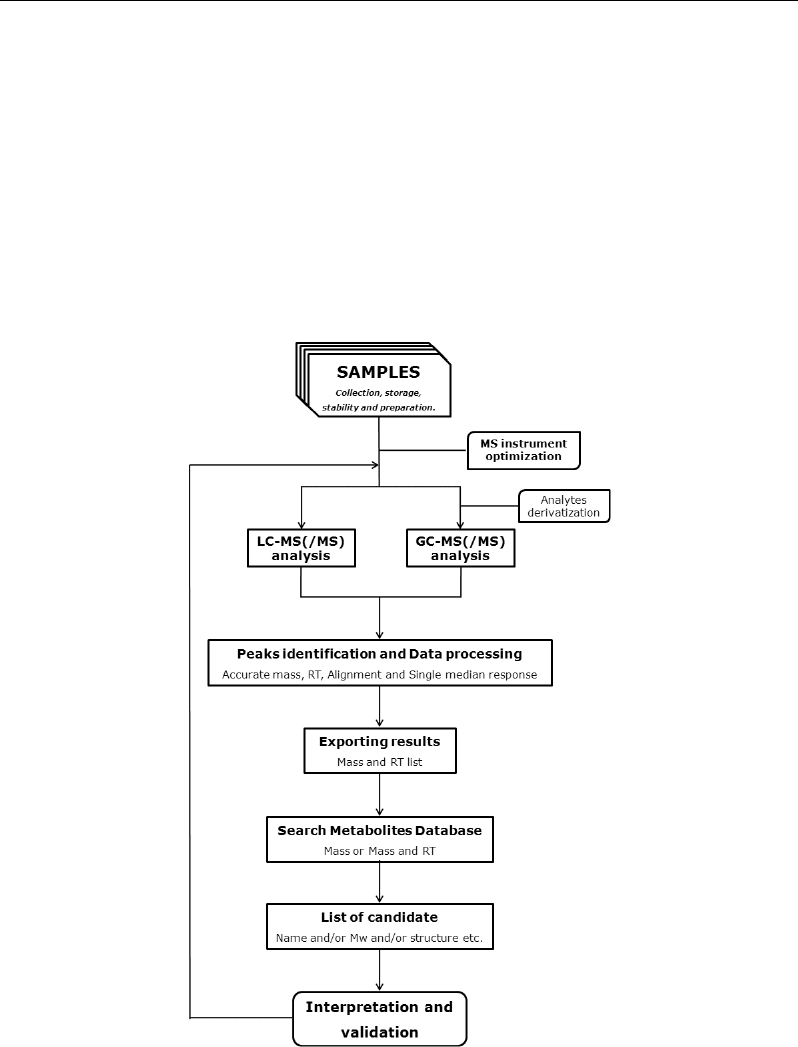
Figure 4. Workflow of a metabolomics GC-MS or LC-MS(/MS) platform.
Ion mobility spectrometry (IMS) was introduced in the ’70s as a low resolution ion-
separation and detection device [218]. IMS is based on the fact that ions with different shapes
travel at different speeds when they are pulled by a weak electric field through a drift cell
filled with a buffer gas. Coupling of IMS to MS results in a 2D, orthogonal separation
technique. IMS was interfaced with ToF-MS in the late ’90s [219], permitting the

Chiara Cavaliere, Eleonora Corradini, Patrizia Foglia et al. 138
simultaneous acquisition of ion mobility spectra and mass spectra in a single run. The
combined LC-IM-MS approach using Q-ToF with electrospray ionization, has been recently
tested for urine metabolome study, and demonstrates the potential for high coverage, high
throughput analysis [220].
Whatever the technological platform (hardware) was chosen, a metabolomic experiment
include, in addition to the experimental design, a series of operational steps as reported, for
example, in Figure4.
Direct Infusion MS and MS/MS
Direct MS (DMS) analyses of complex metabolite mixtures in conjunction with
chemometric data analysis offer a viable solution when high-throughput screening is
mandatory. The high-throughput capacity of DMS fingerprinting of complex mixtures is
similar to that of NMR fingerprinting.
Direct infusion MS (DIMS), or flow injection of complex metabolic extracts without
chromatographic separation via ESI provides a sensitive, high-throughput method for
metabolic fingerprinting. The greater sensitivity compared to NMR makes it a very useful
approach for large-scale screening. Obviously, DI-ESI-MS analysis is much more susceptible
to ionization suppression than LC-ESI-MS, thus it is not usually advocated as a quantitative
method. The utilization of nano-ESI reduces ionization suppression effects due to the
increased ionization efficiency of nano-DIMS, and chip-based nanospray emitters provide a
fully automated platform for high-throughput DIMS metabolite measurements [221]. DIMS is
unable to differentiate among isomeric compounds, however DIMS
n
using ion traps produces
fragment ions that often enable the differentiation of isomeric structures. Recently a rapid
metabolic profiling method using both untargeted and targeted DIMS
3
with a relatively low
resolution linear ion trap mass spectrometer has been shown to yield sufficient precision and
accuracy for application in genetical metabolomics provided that suitable software tools were
developed [222].
FT-ICR-MS is a powerful tool for DIMS because of its very high mass resolution (10
6
)
and mass accuracy (<1 ppm), and it has been successfully applied for metabolite-
fingerprinting studies [223,224]. However, large ion populations influence negatively the
mass-measurement accuracy and limit the dynamic range of FT-ICR when a wide m/z range
is tapped. Narrow m/z ranges are therefore commonly acquired separately to increase the
dynamic range and mass accuracy for metabolic profiling. An optimized strategy, namely
high sensitivity selected ion monitoring (SIM)-stitching approach, is usually followed. Each
wide-scan mass spectrum is recorded as a series of overlapping selected ion monitoring (SIM)
windows that are stitched together using novel algorithms [225]. This, reducing space-charge
effects, increases the dynamic range and maintains high mass accuracy.
Ion suppression during ESI, ion–ion interactions in the detector cell, and thermally-
induced white noise remain major challenges for this approach [226]. Orbitraps may be a
good alternative to the more expensive and higher maintenance FT-ICR mass spectrometers,
with similar resolving powers and mass accuracies of 2–5 ppm.
From Metabolic Profiling to Metabolomics 139
Desorption and Imaging MS
MALDI-MS is a popular analytical technique for protein and peptide analysis. Although
the high throughput nature of MALDI-MS makes it an ideal tool for large-scale metabolomic
studies, its application in the field has been rather limited. Due to the elevated chemical
background signals generated by the matrix in the low molecular-weight region that obscures
the detection of metabolites in the range, MALDI-MS has been confined to the targeted
analysis of high-molecular weight metabolites [227,228]. Recent advancements in laser
desorption techniques include matrices that have minimal background signals in the low-
molecular-weight region (‘‘ionless matrices’’), yet still assisting an efficient
ionization/desorption of the analytes [229,230], and DI-MS from porous silicon chips [231].
These offer exciting new opportunities for the utilization of desorption-ionization in
metabolite screening and fingerprinting; however, the technique is still subject to ion
suppression and yields poor quantitative detection of metabolites [232].
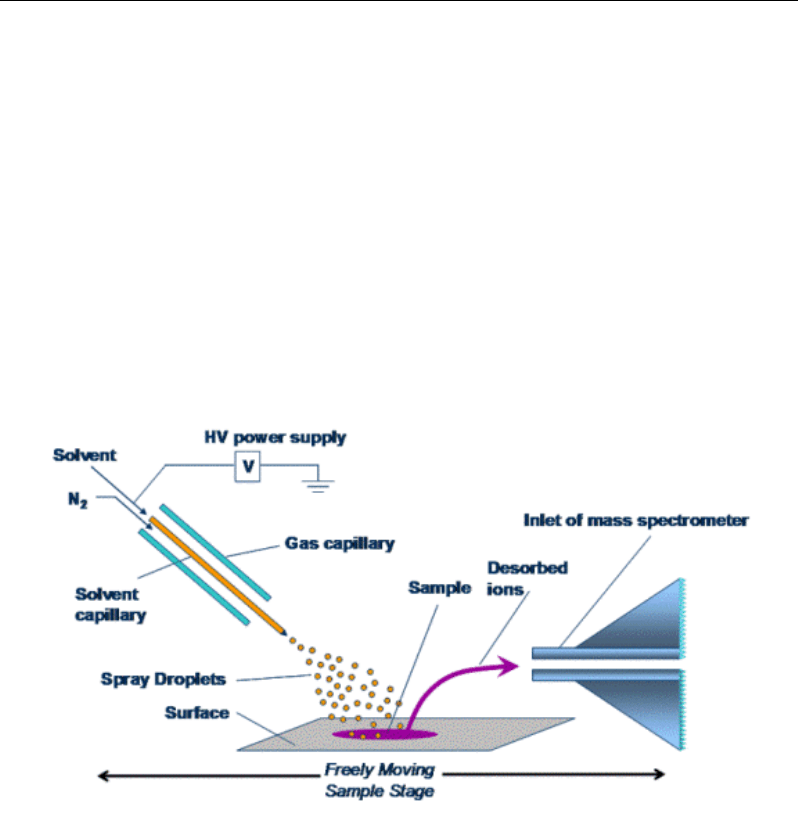
Figure 5a. DESI (desorption electrospray) scheme.
Desorption ESI (DESI) is a ionization technique that combines features of ESI and
desorption ionization to permit analysis directly from a surface with virtually no sample
preparation. An electrospray emitter is used to generate a spray of charged micro droplets that
is directed towards an ambient sample surface. Molecules on the surface are subsequently
desorbed, ionized, desolvated and directed to the MS inlet [233]. Virtually no sample
preparation is required for DESI, thus allowing the direct analysis of tissues or biological
fluids that can be deposited on an inert surface and then analyzed. However, DESI
experimental conditions typically require optimization for each sample type, so time must be
invested initially in optimizing the experimental parameters such as the chemical and physical
nature of the surface and the nature of the spraying solvent [234]. Also the geometry of the
system sample surface- sprayer tip- MS inlet, directly affects the ionization efficiency and
sensitivity. DESI seems to have a higher tolerance to sample-matrix effects than ESI,
however the quantitative precision of DESI, as of other surface ionization techniques, is less
Chiara Cavaliere, Eleonora Corradini, Patrizia Foglia et al. 140
than that of ESI. The application of DESI in metabolomics is relatively new, but its ambient
DI properties as well as its high throughput capacity make it an attractive tool for
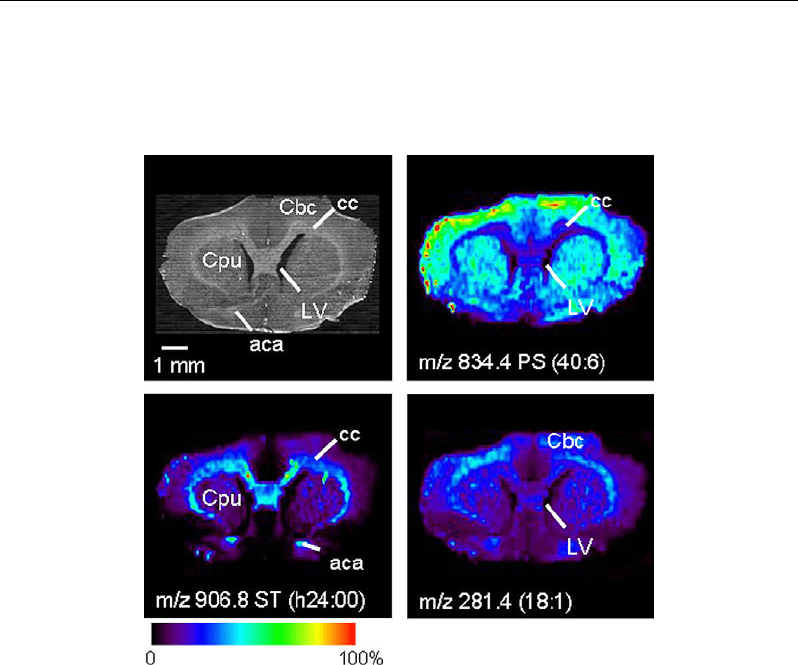
metabolomics. Figure 5a shows a DESI scheme, and Figure 5b the images produced from an
analysis of lipids in a rat brain tissue sample.
Figure 5b. Images produced from an analysis of lipids in a rat brain tissue sample. The first (up left) is
an optical image, and the others are ion images created from desorption electrospray ionization
analysis.
Extractive ESI (EESI) is another new ESI technique that uses two separate sprayers. One
sprayer nebulizes the sample solution that intersects with a second electrospray containing
charged micro-droplets of the ionizing reagent solvent, usually an acidic aqueous methanol.
Analyte molecules are ionized following collision with the reagent micro-droplets and then
mass analyzed. EESI is related to DESI, but was developed for the direct analysis of trace
compounds in the solution phase, especially when complex mixtures are of interest [235].
EESI utilizes two spray sources, eliminating the use of a surface on which the analyte is first
collected. One spray source nebulizes the sample while the other provides charged solvent
droplets. The two spray sources are set at an angle to each other and to the mass spectrometer
inlet so as to introduce the analyte of interest directly to the source [236]. The advantage of
EESI is its ability to analyze complex biological samples, such as urine and serum, directly,
with minimum or no sample preparation for an extended period of time. The direct infusion of
such complex biological samples to conventional ESI ion sources causes an irrecoverable loss
in signal intensity due to the formation of salt adducts, sample carry-over, or cumulative
build-up of non-volatile components in the ion source. The long-term spray stability of
untreated biological samples in EESI is very promising for high throughput metabolomics, as
EESI significantly reduces data-collection interruptions due to frequent ion source cleaning to

From Metabolic Profiling to Metabolomics 141
remove non-volatile accumulations associated with the ESI-DIMS of crude biological
samples [143]. EESI may represents a valid, more sensitive alternative to NMR in
metabolomics; recently, it has been demonstrate that NMR data and EESI mass spectral data
can be cross-validated [237].
Imaging MS (IMS) is an other emerging technology that permits the direct analysis and
determination of the distribution of molecules in tissue sections. Tissues are analyzed intact
and thus spatial localization of molecules within a tissue is preserved [238]. To investigate the
spatial distribution of specific biomolecules, an increasing number of work groups are aiming
to combine the sensitivity and specificity of mass spectrometry with imaging capabilities.
This has included both MALDI and ESI, and has provided fresh impetus to secondary ion
mass spectrometry (SIMS) imaging. The potential of IMS has long been recognized. The first
elemental imaging experiments using SIMS were made in the ’60s [239,240]. However,
widespread bimolecular IMS had to wait for the advent of MALDI and SIMS methodologies
capable of generating ions from biomolecules with sufficient sensitivity. The high mass
capabilities of MALDI enabled it to be readily applied for imaging protein distributions
within tissue sections [241], whereas the principle SIMS application in semi-conductor
research directed its development toward higher spatial resolution. The success of imaging
MALDI has begun to steer SIMS developments toward high mass molecules while retaining
the high spatial resolution capabilities. However, as a rule, MALDI imaging and SIMS
imaging provide spatial information about different classes of biological compounds. MALDI
provides proteomics information, and SIMS that of lipids and other surface active, relatively
low MW species. Hence, MALDI can record the spatial distribution of high mass molecules
using the chemically specific molecular ions; however, typical spatial resolutions are
approximately 25 µm or more (10 µm sources are now available). SIMS is able to provide
high spatial resolution images, sub-micron is routine and 50 nm is commercially available
[242];
however, the molecular ion mass range is much lower than that of MALDI, most
imaging experiments use ions of m/z <500.
MALDI can analyze hundreds of proteins directly from tissue sections, and combining
this with spatial coordinates, allows the spatial distribution of these proteins to be determined
in parallel, without a label and within practical time-scales [243]. The sample preparation,
spatial resolution and sensitivity of the ionization step, are all important parameters that affect
the type of information obtained. Recently, significant progress has been made in each of
these steps for both SIMS and MALDI imaging of biological samples. Mass resolution is an
important feature because it defines the degree of chemical specificity and, through
improving the precision of mass measurement, helps improve mass accuracy. The ultra-high
mass resolution and mass accuracy of the Fourier transform ion cyclotron resonance mass
spectrometer, resolution >100,000 and sub part-per-million mass accuracy, is beginning to be
developed for imaging mass spectrometry experiments [244,245].
The advantage of a SIMS analysis is that tissue sections did not need any further sample
preparation steps: the sample plate can be mounted and the analysis performed. Impressive
biological images of atomic ions and low mass fragments can be obtained directly from
tissue. The sensitivity improvements provided by polyatomic primary ions, added as very thin
films, show sufficient intensity for imaging of intact biomolecules [246-248].
DESI, being an atmospheric pressure (AP) surface ionization technique, can be used as a
chemical IMS. Many lipid species are easily ionized by DESI, making them attractive target
molecules from which to create molecular images of thin tissue sections. DESI-MS has been

Chiara Cavaliere, Eleonora Corradini, Patrizia Foglia et al. 142
used to construct chemical images of tissue sections of mouse pancreas, rat brain, and
metastatic human liver adenocarcinoma, as well as whole tissue analysis of adipose tissue
surrounding a chicken heart giving strong signals from the major lipid components of
biological membranes [249-253]. Theoretically, DESI may be coupled with all the MS and
MS/MS analyzer and, although the IMS application of DESI has been limited to
phospholipids, the technique seems to have the potential for a wider metabolite profiling.
AP-IMS techniques are particularly attractive because, in principle, working at AP also
enables the study of live specimens. Laser ablation ESI (LAESI) [254,255] is a new AP-IMS
technique. LAESI IMS is realized as a combination of lateral imaging and depth profiling by
tissue ablation with single laser pulses of ca. 2.94 μm wavelength. The O-H vibrations of the
native water molecules in the tissue samples readily absorbed the laser pulse energy leading
to ablation, i.e., the ejection of a microscopic volume of the sample in the form of neutral
particulates and/or molecules. This plume was then intercepted by an electrospray, and the
ablated material was efficiently post-ionized. Tandem MS was performed on numerous ions
to help with metabolite structure assign. A recent, novel development of LAESI was the
combination of lateral imaging and molecular depth profiling capabilities, which enabled a
truly 3D metabolite distribution imaging. Although 3D ambient imaging with LAESI has
been proven feasible in plant tissues, further improvements are needed in spatial resolution
[255].
Data Handling
A great drawback of all “omics” sciences where a large number of qualitative and
quantitative data can be generated per single run is the problem of data analysis. Hence, there
is a demand for computational tools to handle and interpret the large amounts of data. For
metabolomics to be successful, raw analytical data must be converted into metabolites
(named chemicals) and their concentrations, or variation of concentration, data that can be
usefully interpreted for biological research.
The data-processing techniques for the deconvolution of chromatographic peaks, library
search, and GC retention index developed originally by Kovats [256] have been in use since
the ’70s to help identification of peaks recorded by GC-MS. [257] Metabolite profiling by
chromatography-MS and statistical analysis still relies on efficient data-processing
procedures, and minimum reporting requirements have recently been suggested [258,259]. In
all published metabolomic studies based on GC-MS, the starting point of the data analysis has
been the deconvolution of chromatograms, followed by peak matching, and then
identification of the differences between samples. Multivariate methods have been developed
and used to clarify chromatographic and spectral profiles from overlapping chromatographic
peaks obtained using various types of hyphenated chromatography systems. The multivariate
curve resolution methods can be divided into iterative, non-iterative, and hybrid approaches,
and all present both advantages and disadvantages [134]. Software programs have been
developed capable of spectral filtering (noise elimination), peak detection finding the peaks
corresponding to the same compounds, m/z alignment (aiming at matching the corresponding
peaks across multiple sample runs) and normalization (adjusting the intensities within each
sample run by reducing the systematic error) [260-264]. Software packages have augmented
their capability during the years, and many can be downloaded from the Internet.

From Metabolic Profiling to Metabolomics 143
After data alignment and correction of retention times, a data matrix can be generated
from the peak lists as output for subsequent multivariate statistical analysis, including
supervised classification for fingerprinting and principal component analysis for data
visualization [153]. Visualization of complex mass spectrometric data sets is becoming
increasingly important in metabolomics. In a recent paper a versatile tool suitable for many
frequently occurring tasks handling LC-MS data is described [265]. Depending on the task at
hand, different views may be used: single spectra visualized as a plot of intensity against
mass-to-charge ratio (1D view); LC-MS maps displayed in a 2D view from a bird’s-eye
perspective with color-coded intensities; selected regions of LC-MS maps displayed in a 3D
view.
The chemical identification of metabolites is fundamental for the extraction of biological
context from the data. It is not easy to identify a metabolite in a metabolome, especially low
level metabolites which are at or slightly above the noise level or are masked by other
metabolites. An estimated total number of possible metabolites ranges from 200,000 to
1,000,000 [4,266]. A number of strategies are being tried out to assist in the chemical
identification of the unknowns, including the development of metabolite-specific mass
spectral libraries and databases [267-270]. The basis on which metabolites are identified
varies among the metabolomics community. In an effort to standardize the reporting and
interpretation of metadata, the Metabolomics Standard Initiative (MSI) [271] has formed five
working groups that follow the general workflow model in metabolomics: biological context;
chemical analysis; data processing; ontology; and data exchange.
Two types of identification are possible: putative or preliminary identification and
definite identification. A global metabolomic study involves the search for all metabolites in a
biological specimen. If the search is for known metabolites, the identification involves
comparing the experimental data with that of pure standards. Experimentally determined
accurate mass or electron-impact mass spectrum are typically applied for putative
identification. In LC-MS and DIMS the accurate mass is used to define molecular formulae
from which suitable metabolites can be derived by searching electronic resources. However,
isomers have the same accurate mass and therefore require a separate, orthogonal property for
definite identification of all potential isomers. Most metabolite identifications reported are
typically non-novel as they have been previously characterized, identified, and reported at a
rigorous level in literature. Thus, non-novel metabolites not being identified for the first time
are typically identified through the co-characterization with authentic chemical standards. The
Chemical Analysis Working Group has recently proposed a guideline for the identification of
non-novel metabolites [258], in which a minimum of two independent and orthogonal data
relative to an authentic standard compound, typically retention time or index and
fragmentation mass spectrum analyzed under identical experimental conditions, are
considered necessary for metabolite identification.
However, if the metabolites are not known, metabolite identification is much more
difficult. This effort would involve multiple chromatographic separations using different
column chemistries and mobile phases. It also would require high resolution MS and MS/MS
for accurate mass analysis, statistical analysis software for data mining, metabolite databases,
and access to an index of hundreds of pure compounds for confirmation. Databases are still at
an early level of development, and techniques, such as chemical-formula determination via
accurate mass, often serve only to narrow the potential search field rather than provide an
unambiguous formula. Biomarker identification therefore remains a potentially time-

Chiara Cavaliere, Eleonora Corradini, Patrizia Foglia et al. 144
consuming process and a limitation for this technique. Some freely and commercially
available web-resources for MS-based metabolomics are mainly reported in references
[261,272] and also in the literature quoted in this section.
Future Perspectives
LC-MS is certainly going to become a key technology for the provision of global
metabolomics. Major advances in available technologies and a totally new approach to
analysis are essential before a true metabolomics, not achievable with the current state of
development in analytical science, can be performed. This will include developments in all
steps of metabolite analysis, from sampling, sample storage and preparation, to data
acquisition, storage and processing, coupled with a greater understanding and application of
bioinformatics.
Future developments in exhaustive mining of the metabolome will no doubt evolve
through improvements in chromatographic technologies and sensitive MS instruments with a
high capability for mass accuracy. Continued improvements in miniaturized MS, multi-
dimensional chromatography, and multiplexed MS approaches will offer new opportunities in
metabolomics. An increase in the number of chemically identifiable metabolites is a
fundamental key to the interpretation and understanding of the biological context of
metabolomics experiments. High throughput techniques, such as DESI and EESI, are very
attractive and will certainly undergo major improvements in sensitivity, dynamic range, and
high resolution MS/MS coupling. Finally, chemical imaging techniques based on MS are very
exciting and, although not a novelty, still in their infancy. In vivo analysis of intact samples is
an attractive proposition, and in this context, NMR-based technologies have a distinct
advantage over instruments based on MS. But every gap may be overcome.
References
[1] van der Werf, MJ; Jellema, RH; Hankemeier, T. Towards replacing closed with open
target selection strategies. J. Ind. Microbiol. Biotechnol., 2005 32, 234-252.
[2] van der Werf, MJ. Microbial metabolomics: replacing trial-and-error by the unbiased
selection and ranking of targets. Trends Biotechnol., 2005 23, 11-16.
[3] Sumner, LW; Mendes, P; Dixon, RA. Plant metabolomics: large-scale phytochemistry
in the functional genomics era. Phytochemistry, 2003 62, 817-36.
[4] Fiehn, O; Kopka, J; Dormann, P; Altmann, T; Trethewey, RN; Willmitzer, L.
Metabolite profiling for plant functional genomics. Nat. Biotechnol., 2000 18, 1157-
1161.
[5] Raamsdonk, LM; Teusink, B; Broadhurst, D; Zhang, N; Hayes, A; Walsh, MC; Berden,
JA; Brindle, KM; Kell, DB; Rowland, JJ; Westerhoff, HV; van Dam, K; Oliver, SG. A
functional genomics strategy that uses metabolome data to reveal the phenotype of
silent mutations. Nat. Biotechnol., 2001 19, 45-50.
[6] Clish, CB; Davidov, E; Oresic, M; Plasterer, TN; Lavine, G; Londo, T; Meys, M; Snell,
P; Stochaj, W; Adourian, A; Zhang, X; Morel, N; Neumann, E; Verheij, E; Vogels, JT;

From Metabolic Profiling to Metabolomics 145
Havekes, LM; Afeyan, N; Regnier,F; van der Greef, J; Naylor, S. Integrative biological
analysis of the APOE3-leiden transgenic mouse. Omics, 2004 8, 3-13.
[7] Goodacre, R; Vaidyanathan, S; Dunn, WB; Harrigan, GG; Kell, DB. Metabolomics by
numbers: acquiring and understanding global metabolite data. Trends Biotechnol., 2004
22, 245-252.
[8] Brindle, JT; Antti, H; Holmes, E; Tranter, G; Nicholson, JK; Bethell, HW; Clarke, S;
Schofield, PM; McKilligin, E; Mosedale, DE; Grainger, DJ. Rapid and noninvasive
diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based
metabonomics. Nat. Med., 2002 8, 1439-1444.
[9] Sabatine, MS; Liu, E; Morrow, DA; Heller, E; McCarroll, R; Wiegand, R; Berriz, GF;
Roth, FP; Gerszten, RE. Metabolomic identification of novel biomarkers of myocardial
ischemia. Circulation, 2005 112, 3868-3875.
[10] Kawashima, H; Oguchi, M; Ioi, H; Amaha, M; Yamanaka, G; Kashiwagi, Y;
Takekuma, K; Yamazaki, Y; Hoshika, A; Watanabe, Y. Primary biomarkers in cerebral
spinal fluid obtained from patients with influenza-associated encephalopathy analyzed
by metabolomics. Int J. Neurosci., 2006 116, 927-936.
[11] [11] Ippolito, JE; Xu, J; Jain, S; Moulder, K; Mennerick, S; Crowley, JR; Townsend,
RR; Gordon, JI. An integrated functional genomics and metabolomics approach for
defining poor prognosis in human neuroendocrine cancers. Proc. Natl. Acad. Sci. 2005
102, 9901-9906.
[12] Ryan, D; Robards, K. Metabolomics: The greatest omics of them all? Anal. Chem.,
2006 78, 7954-7958.
[13] Oliver, SG; Winson, MK; Kell, DB; Baganz, F. Systematic functional analysis of the
yeast genome. Trends Biotechnol., 1998 16, 373-378.
[14] Goodacre, R. Making sense of the metabolome using evolutionary computation: seeing
the wood with the trees. J. Exp. Bot., 2005 56, 245-254.
[15] Fiehn, O. Combining genomics, metabolome analysis, and biochemical modelling to
understand metabolic networks. Comp. Funct. Genom., 2001 2, 155-168.
[16] Nicholson, JK; Lindon, JC; Holmes, E. “Metabomics": understanding the metabolic
responses of living systems to pathophysiological stimuli via multivariate statistical
analysis of biological NMR spectroscopic data. Xenobiotica, 1999 29, 1181-1189.
[17] Fiehn, O. Metabolomics - the link between genotypes and phenotypes. Plant Mol. Biol.,
2002 48, 155-71.
[18] Lind
on, JC; Holmes, E; Nicholson, JK. So what's the deal with metabonomics? Anal.
Chem., 2003 75, 384A-391A.
[19] MacKenzie, DA; Defernez, M; Dunn, WB; Brown, M; Fuller, LJ; de Herrera, S;
Guenther, A; James, SA; Eagles, J; Philo, M; Goodacre, R; Roberts, IN. Relatedness of
medically important strains of Saccharomyces cerevisiae as revealed by phylogenetics
and metabolomics. Yeast, 2008 25, 501-512.
[20] Smedsgaard, J; Nielsen, J. Metabolite profiling of fungi and yeast: from phenotype to
metabolome by MS and informatics. J. Exp. Bot., 2005 56, 273-286.
[21] Allwood, JW; Ellis, DI; Heald, JK; Goodacre, R; Mur, LAJ. Metabolomic approaches
reveal that phosphatidic and phosphatidyl glycerol phospholipids are major
discriminatory non-polar metabolites in responses by Brachypodium distachyon to
challenge by Magnaporthe grisea. Plant J., 2006 46, 351-368.

Chiara Cavaliere, Eleonora Corradini, Patrizia Foglia et al. 146
[22] Hall, RD. Plant metabolomics: from holistic hope, to hype, to hot topic. New Phytol.,
2006 169, 453-468.
[23] Kell, DB. Systems biology, metabolic modelling and metabolomics in drug discovery
and development. Drug Discov. Today., 2006 11, 1085-1092.
[24] Dunn, WB; Broadhurst, DI; Deepak, SM; Buch, MH; McDowell, G; Spasic, I; Ellis, D;
Brooks, N; Kell, DB; Neyses, L. Serum metabolomics reveals many novel metabolic
markers of heart failure, including pseudouridine and 2-oxoglutarate. Metabolomics,
2007 3, 413-426.
[25] Kenny, LC; Broadhurst, D; Brown, M; Dunn, WB; Redman, CWG; Kell DB; Baker,
PN. Detection and identification of novel metabolomic biomarkers in preeclampsia.
Reprod. Sci., 2008 15, 591-597.
[26] Lindon, JC; Holmes, E; Nicholson, JK. Metabonomics in pharmaceutical R & D. FEBS
J., 2007 274, 1140-1151.
[27] Tanaka, Y; Higashi, T; Rakwal, R; Wakida, S-Ii; Iwahashi, H. Quantitative analysis of
sulfur-related metabolites during cadmium stress response in yeast by capillary
electrophoresis-mass spectrometry. J. Pharm. Biomed. Anal., 2007 44, 608-613.
[28] Viant, MR. Metabolomics of aquatic organisms: the new 'omics' on the block. Mar.
Ecol.-Prog. Ser., 2007 332, 301-306.
[29] Herrgard, MJ; Swainston, N; Dobson, P; Dunn, WB; Arga, KY; Arvas, M; Bluthgen,
N; Borger, S; Costenoble, R; Heinemann, M; Hucka, M; Le Novere, N; Li, P;
Liebermeister, W; Mo, ML; Oliveira, AP; Petranovic, D; Pettifer, S; Simeonidis, E;
Smallbone, K; Spasic, I; Weichart, D; Brent, R; Broomhead, DS; Westerhoff, HV;
Kirdar, B; Penttila, M; Klipp, E; Palsson, BO; Sauer, U; Oliver, SG; Mendes, P;
Nielsen, J; Kell, DB. A consensus yeast metabolic network reconstruction obtained
from a community approach to systems biology. Nat. Biotechnol., 2008 26, 1155-1160.
[30] Wishart, DS; Knox, C; Guo, AC; Eisner, R; Young, N; Gautam, B; Hau, DD;
Psychogios, N; Dong, E; Bouatra, S; Mandal, R; Sinelnikov, I; Xia, J; Jia, L; Cruz, JA;
Lim, E; Sobsey, CA; Shrivastava, S; Huang, P; Liu, P; Fang, L; Peng, J; Fradette, R;
Cheng, D; Tzur, D; Clements, M; Lewis, A; De Souza, A; Zuniga, A; Dawe, M; Xiong,
Y; Clive, D; Greiner, R; Nazyrova, A; Shaykhutdinov, R; Li, L; Vogel, HJ; Forsythe, I.
HMDB: a knowledge-base for the human metabolome. Nucleic Acids Research, 2009
37, D603-D610.
[31] Brown, M; Dunn, WB; Ellis, DI; Goodacre, R; Handl, J; Knowles, JD; O’Hagan, S;
Spasic, I; Kell, DB. A metabolome pipeline: from concept to data to knowledge.
Metabolomics, 2005 1, 39-51.
[32] Dunn, WB. Current trends and future requirements for the mass spectrometric
investigation of microbial, mammalian and plant metabolomes. Phys. Biol., 2008 5, 1-
24.
[33] Lu, W; Bennett BD; Rabinowitz, JD. Analytical strategies for LC-MS-based targeted
metabolomics. J. Chromatogr. B, 2008 871, 236-242.
[34] Griffin, JL. Metabonomics: NMR spectroscopy and pattern recognition analysis of
body fluids and tissues for characterisation of xenobiotic toxicity and disease diagnosis.
Curr. Opin. Chem. Biol., 2003 7, 648-654.
[35] Want, EJ; N
ordstrom, A; Morita, H; Siuzdak, G. From Exogenous to Endogenous: The
Inevitable Imprint of Mass Spectrometry in Metabolomics. J. Proteome Res., 2007 6,
459-468.
