Accardi L., Freudenberg W., Obya M. (Eds.) Quantum Bio-informatics IV: From Quantum Information to Bio-informatics
Подождите немного. Документ загружается.


20
4. Construction Cis-module
Datbase
4.1. Data sources
and
Data Collection (Fig4-1)
In this work, the database responsible for five organic species (Homo sapiens,
Mus musculus, Rattus norvegicus, Dorosophila melanogaster and
Saccharomyces serevisiae) was constructed. To collect cis-element information,
we used DDBJ (http://www.ddbj.nig.ac.jpl) which
is
one
of
three members
of
International Nucleotide Sequence Database Collaboration (INSDC), and this
database stores information assured by biochemical experiments.
At the same time, we extracted genome data from Ensembl database
(http://www.ensembl.org/index.html).whichcontainedlocationofgeneloci. By
using Ensembl data, we could bring together data.
4.2. Identification
oj
CDS location on Genome (Fig4-2)
We extracted coding sequences on genome (ensembVCDSs) from genome
sequence data. And then, CDSs
of
DDBJ entries (DDBJ/CDSs) were extracted
from DDBJ data, and compared with ensmebl/CDSs with SSEARCH program.
This process allowed us
to
identify locations
of
records on genome.
4.3. Extraction
and
Comparison upstream region (Fig4-3, 4-4)
Upstream region
of
CDSs
of
DDBJ records (USRlDDBJ) and that
of
ensembl
genome data (USRlensembl) were extracted. And, USRlDDBJ were compared
with corresponding USRlensembl, leading
to
identification
of
cis-element
location on genome. Through this process, we get cis-module entry which
is
cis-module information per gene.
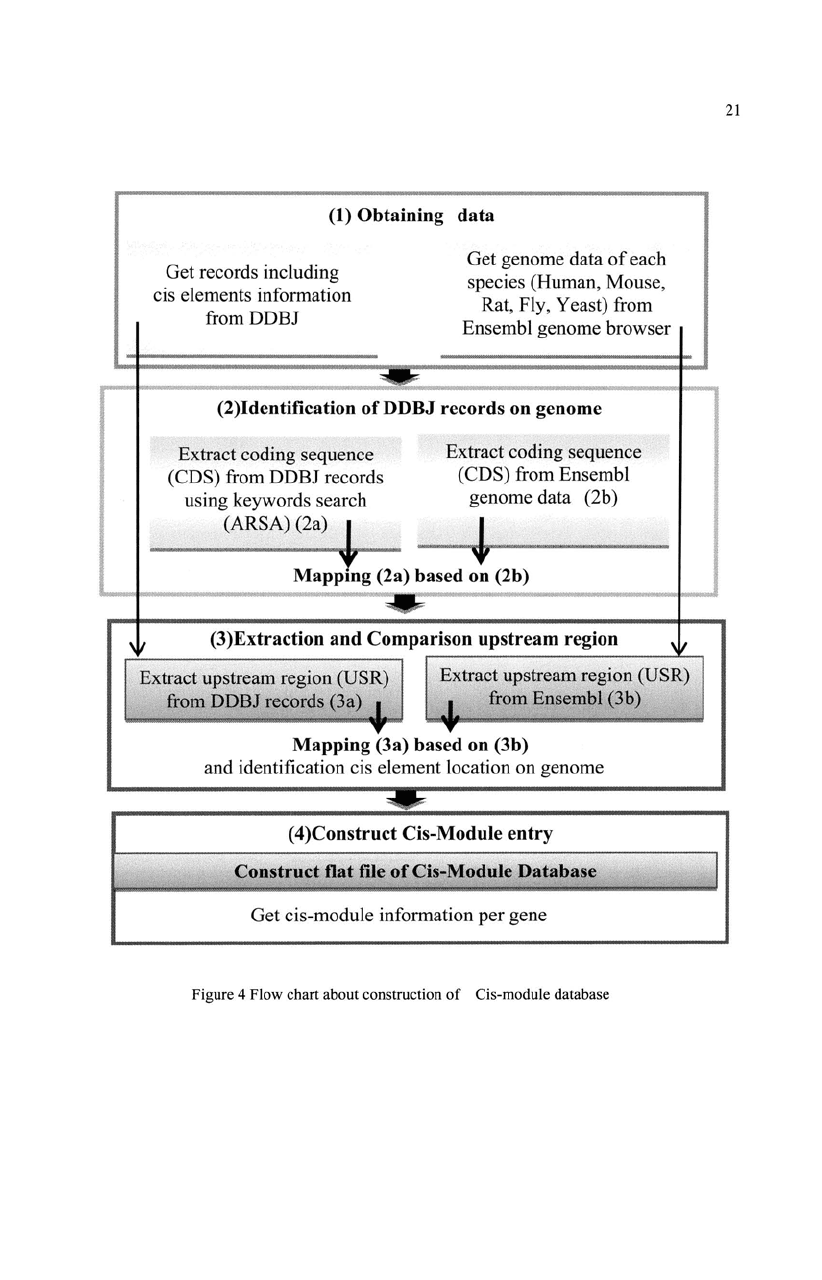
(1) Obtaining data
Get records including
cis elements information
f
romDDBJ
Get genome data
of
each
species (Human, Mouse,
Rat, Fly, Yeast) from
Ensembl genome browser
(2)Identification
of
DDBJ records
on
genome
(CDS) from DDBJ records (CDS) from EnsembI
using keywords search genome data (2b)
(A~
~~:
2:
~
)
+
~
H
~
•.
.
~
i-<l
:h
"'
A~
'
~
~
,"-
""
"'
_>"
----"
.,
/
::'
:
';
~
oai,...;
'
,·-
-
',-.
,-
'~
,
~
~~~
~~C1£
Mappmg (2a) based on (2b)
Mapping (3a) based on (3b)
and identification cis element location on genome
Get cis-module information per gene
Figure 4 Flow chart about construction
of
Cis-module database
21
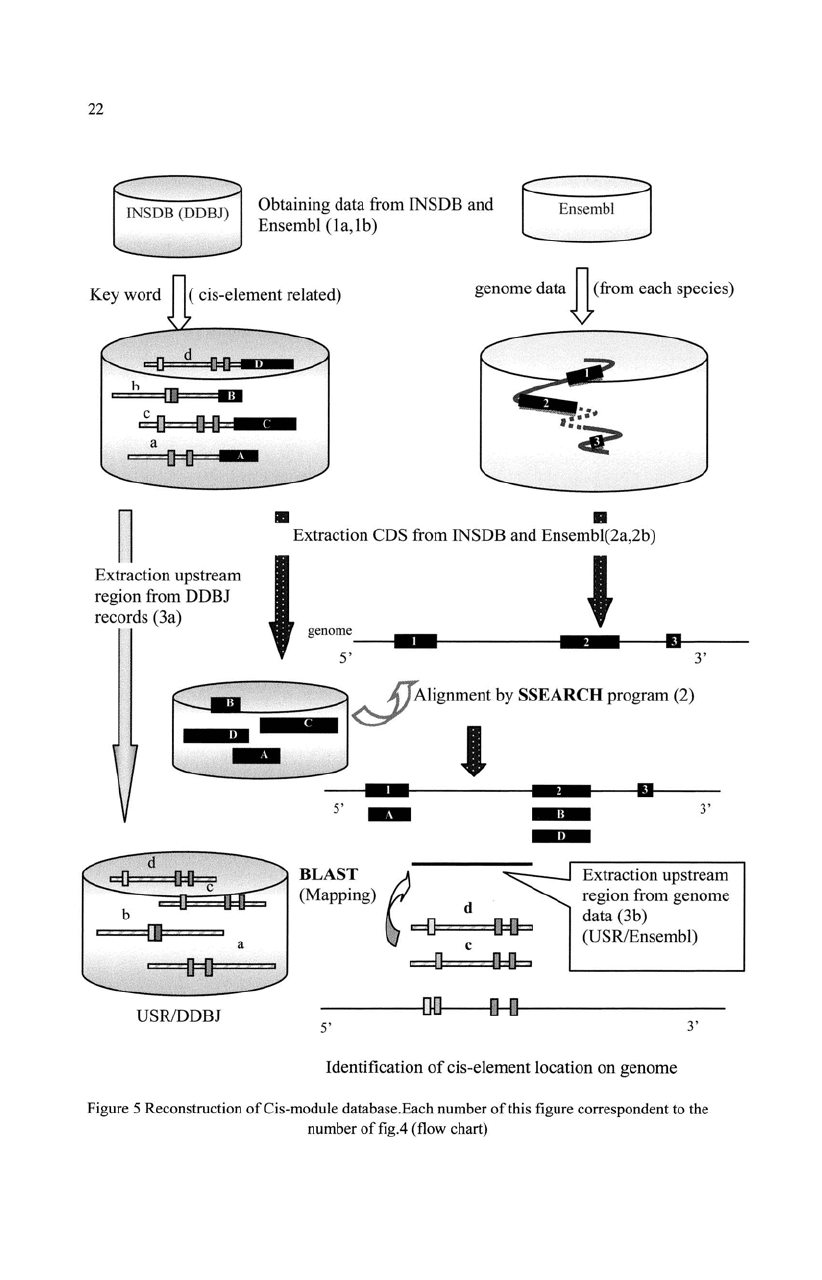
22
Extraction upstream
region from DDBJ
records (3a)
USRlDDBJ
genome data
D(from
each species)
• •
Extraction CDS from INSDB and Ensembl(2a,2b)
-
5'
-
BLAST (
(Mapping)
c:::{I
d
c
II
-
3'
-
Extraction upstream
region from genome
data (3b)
(USRlEnsembl)
~
5'
3'
Identification
of
cis-element location on genome
Figure 5 Reconstruction
of
Cis-module database. Each number
of
this figure correspondent to the
number
offig.4
(flow chart)
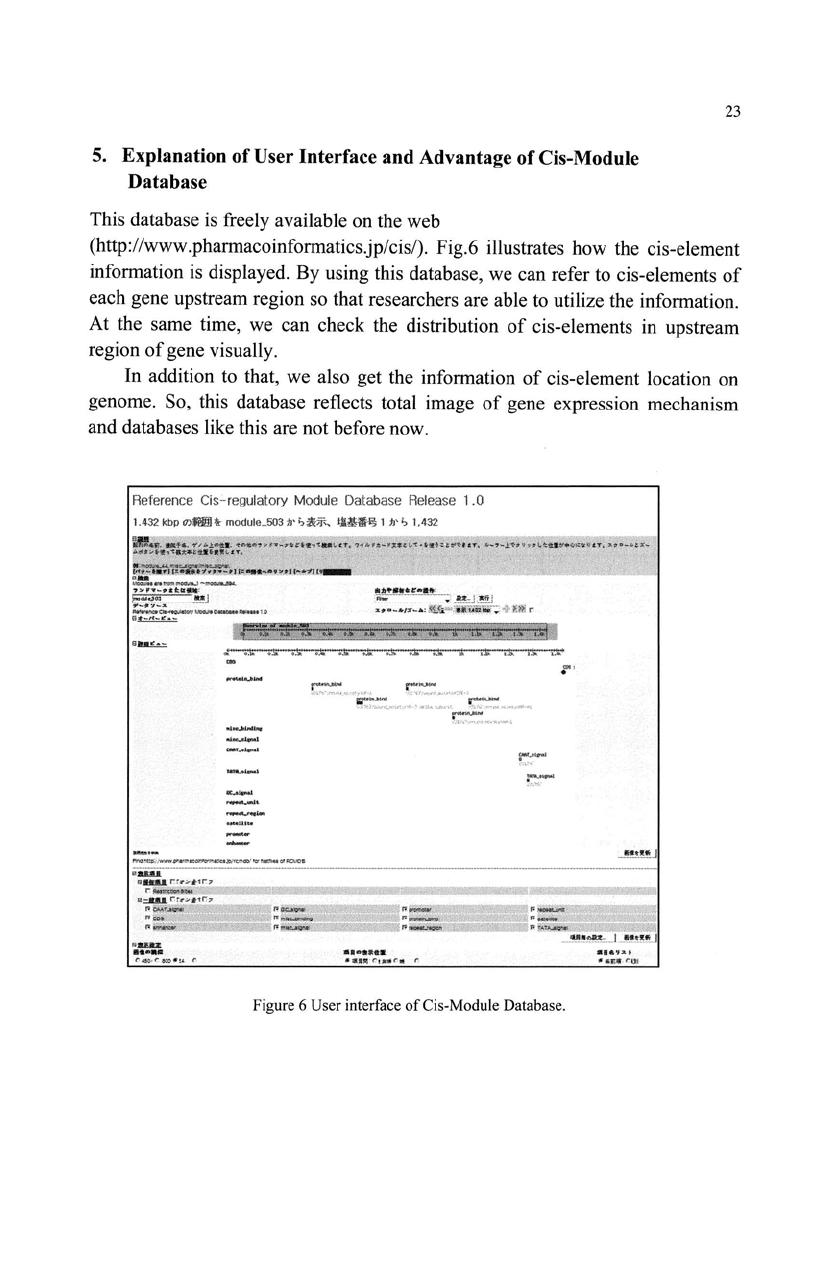
5.
Explanation
of
User Interface and Advantage
of
Cis-Module
Database
23
This database
is
freely available on the web
(http://www.pharmacoinformatics.jp/cis/). Fig.6 illustrates how the cis-element
information
is
displayed. By using this database, we can refer
to
ci
s-elements
of
each gene upstream region so that researchers are able
to
utilize the information.
At the same time, we can
ch
eck the distribution
of
cis-elements in upstream
region
of
gene visually.
In
addition
to
that, we also get the information
of
cis-element location on
genome.
So
, this database reflects total image
of
gene expression mechanism
and databases like this are not before now.
Fieference
C
i
s~r
egul
atory
Module D
at
abase
Fie
lease 1.0
Ilk • •
\:
.
SI:
-
-
--
Figure 6 User interface
ofeis-
Module Database.
24
6.
Application
of
Cis-module
Database
6.1.
Summary
of
Co-occurrence
ofCEs
and
TFsfrom
Cis-module
Database
A gene expression
is
regulated by sets
of
cis-elements and transcriptional factors.
Therefore, we extracted entries that have more than two TF information from
Cis-Module Database (CMDB) to know TFs working together when some gene
express. In table
1,
when the gene (module_7) changes the expression, TF1, TF3
and TF6 work for the change together. So we could get co-occurrence
of
CEs
and TFs from CMDB. By this summary, we could know which TFs and CEs
involved in a gene expression.
Table I : The example
of
co-occurrence information about transcriptional factors (TFs).
TFI TF2
TF3
... TF6 ...
module 7
~¥,~~S~~
r";~AF,~~Y0
:
~~\?~~~~i
;'~
...
0
0
module
431
0
0
6.2. Prediction
of
gene network
for
caloric restriction
Rat
Recently, relationship between anti-aging and calorie control are suggesting. As
the reason for that caloric restriction rodents have longer life.
So
we performed
below process (Fig.7)
to
identify common transcriptional factors, which regulate
differentially-expressed genes under calorie restricted condition.
It
is
expected
that they are involved in a key role for anti-aging.
(1)
Data
analysis
(about
Microarray
data)
We used rat micro array data, which has 31099 genes proved and was kindly
provided by Dr. Higami. In the present study, 3 groups were prepared:
nontransgenic male Wi star rats ad libitum intake (AL), calorie restricted (CR)
rats and heterotransgenic rats (tgl-:
TO). Each group had 4 rats. We targeted at
AL and CR rat to analysis to specify which genes change the expression under
the caloric restricted condition.
AL group rats continued
to
receive food at libtium, and CR group rats were
provided with
70%
of
the mean daily intake
of
AL average. After the
comparison
of
AL data and CR data, differentially expressed 54 genes were
identified.
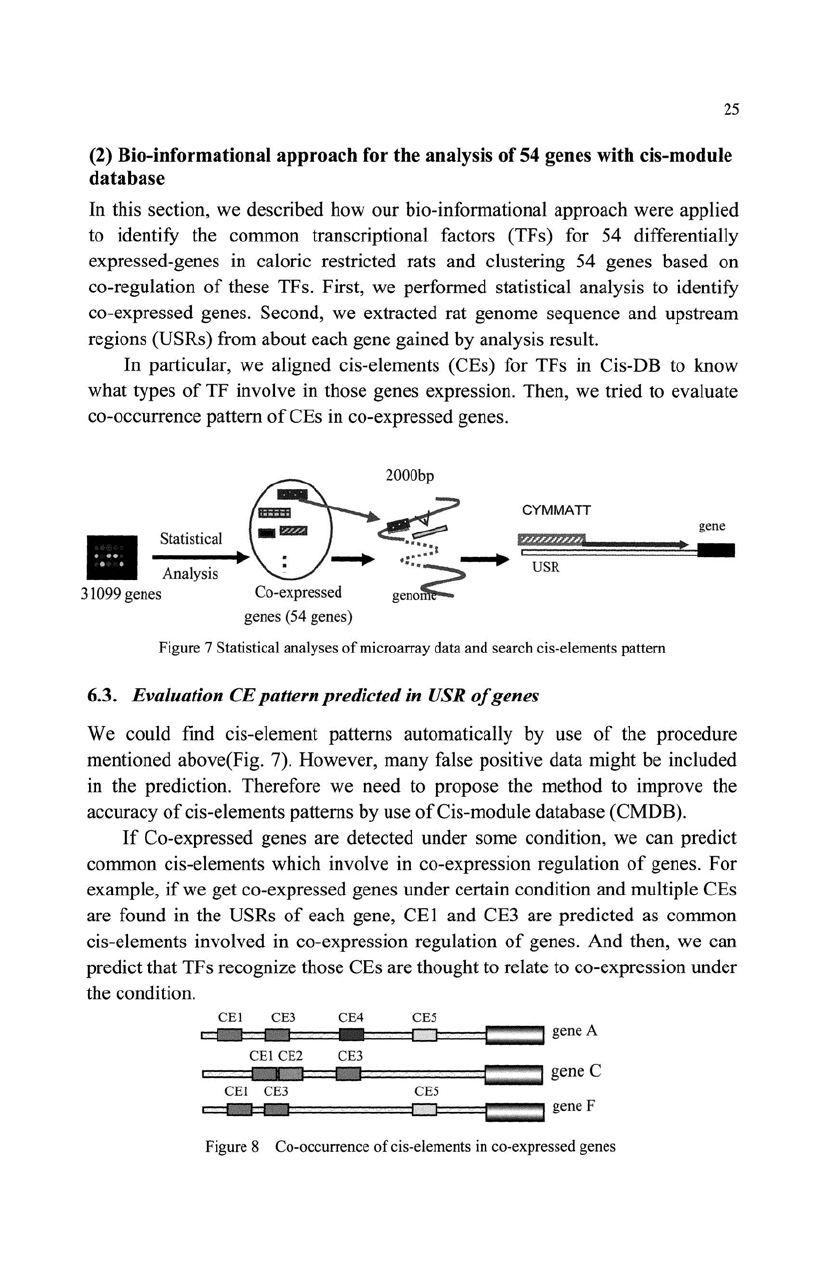
25
(2) Bio-informational
approach
for
the
analysis
of
54 genes with cis-module
database
In this section, we described how our bio-informational approach were applied
to identify the common transcriptional factors (TFs) for 54 differentially
expressed-genes in caloric restricted rats and clustering
54
genes based on
co-regulation
of
these TFs. First, we performed statistical analysis to identify
co-expressed genes. Second, we extracted rat genome sequence and upstream
regions (USRs) from about each gene gained by analysis result.
In particular, we aligned cis-elements (CEs) for TFs in Cis-DB
to
know
what types
of
TF involve in those genes expression. Then, we tried to evaluate
co-occurrence pattern
of
CEs in co-expressed genes.
~~..
~~CY~M~~~A~T~T~~~~~ge.nlle
---~
..
~
:
--..
.:;:"~
-+
t:;;
USR
Analysis·
-:::>
Statistical
31099 genes Co-expressed
geno~
genes (54 genes)
Figure 7 Statistical analyses
of
microarray data and search cis-elements pattern
6.3. Evaluation CE pattern predicted in USR
of
genes
We could find cis-element patterns automatically by use
of
the procedure
mentioned above(Fig.
7).
However, many false positive data might be included
in the prediction. Therefore we need to propose the method
to
improve the
accuracy
of
cis-elements patterns by use
of
Cis-module database (CMDB).
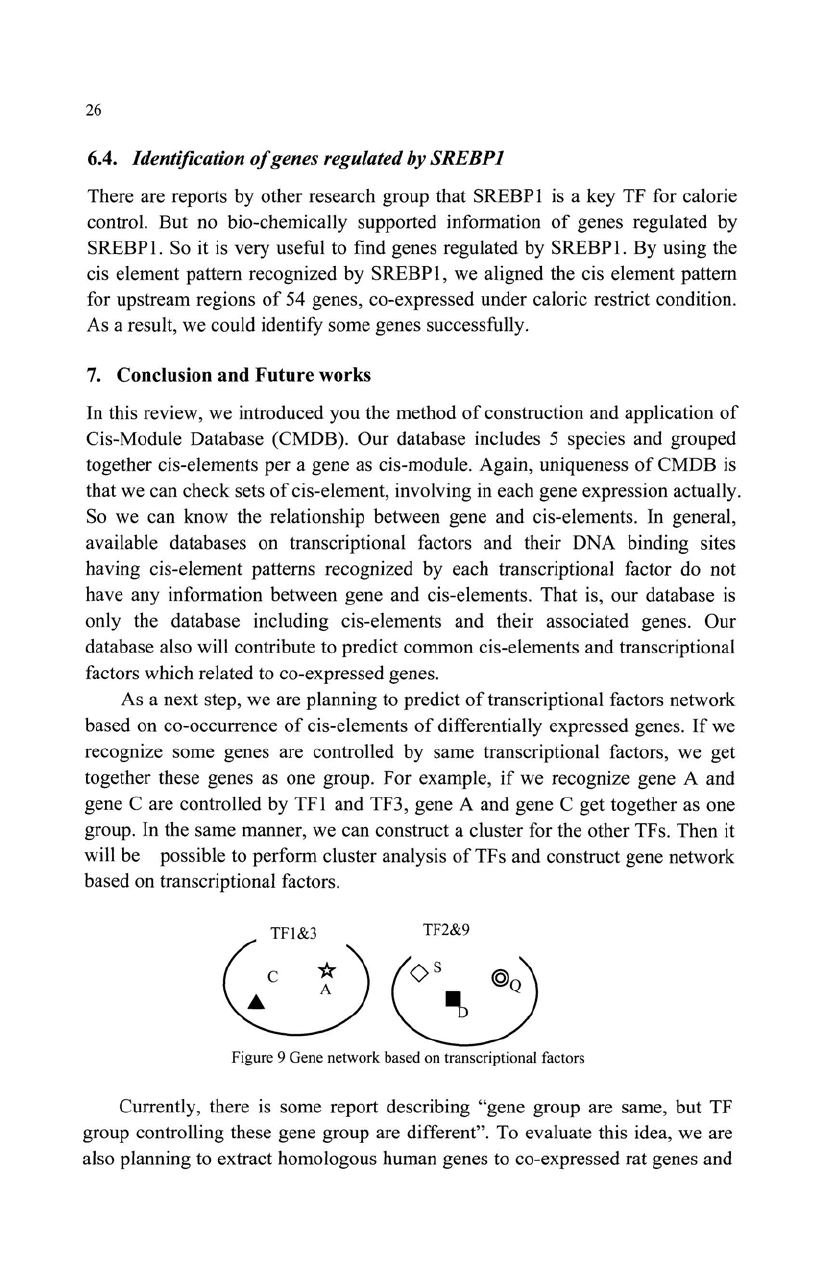
If
Co-expressed genes are detected under some condition, we can predict
common cis-elements which involve in co-expression regulation
of
genes. For
example,
if
we get co-expressed genes under certain condition and multiple CEs
are found in the
USRs
of
each gene, CE 1 and CE3 are predicted
as
common
cis-elements involved in co-expression regulation
of
genes. And then, we can
predict that TFs recognize those CEs are thought to relate to co-expression under
the condition.
CE I
CE3
CE4
CES
c~::I!D=
..
:::1
..
I!iil~=:I_
•.
===I~
gene A
CEI CE2
CE3
-
c::::J gene C
CEI
CE3 CE5
~c·=·
_lI=:Ii!II!1:=====:::::[~I::':=··
.:=
..
:=
...
1'l"m
..••.
,.""
••••
I'!I!J.,
gene F
Figure 8 Co-occurrence
of
cis-elements in co-expressed genes
26
6.4. Identification
of
genes regulated by
SREBPI
There are reports by other research group that SREBP 1
is
a key TF for calorie
control. But no bio-chemically supported information
of
genes regulated by
SREBP
1.
So
it
is
very useful to find genes regulated by SREBP
1.
By using the
cis element pattern recognized by
SREBP
1,
we aligned the cis element pattern
for upstream regions
of
54
genes, co-expressed under caloric restrict condition.
As a result, we could identify some genes successfully.
7.
Conclusion
and
Future
works
In this review, we introduced you the method
of
construction and application
of
Cis-Module Database (CMDB). Our database includes 5 species and grouped
together cis-elements per a gene
as
cis-module. Again, uniqueness
of
CMDB
is
that we can check sets
of
cis-element, involving in each gene expression actually.
So
we can know the relationship between gene and cis-elements. In general,
available databases on transcriptional factors and their DNA binding sites
having cis-element patterns recognized by each transcriptional factor
do
not
have any information between gene and cis-elements. That is, our database
is
only the database including cis-elements and their associated genes. Our
database also will contribute
to
predict common cis-elements and transcriptional
factors which related to co-expressed genes.
As a next step, we are planning to predict
of
transcriptional factors network
based on co-occurrence
of
cis-elements
of
differentially expressed genes.
If
we
recognize some genes are controlled by same transcriptional factors, we get
together these genes
as
one group. For example,
if
we recognize gene A and
gene C are controlled by TFI and TF3, gene A and gene C get together
as
one
group. In the same manner, we can construct a cluster for the other TFs. Then it
will be possible
to
perform cluster analysis
of
TFs and construct gene network
based on transcriptional factors.
08
Figure 9 Gene network based on transcriptional factors
Currently, there
is
some report describing "gene group are same, but
TF
group controlling these gene group are different". To evaluate this idea, we are
also planning
to
extract homologous human genes
to
co-expressed rat genes and

27
construct gene network
of
Human (For example, gene D and gene
D'
have
homology). And then, we intend to compare gene network for other species.
It
may indicate that transcriptional regulatory mechanisms are variable among
species. We believe that our result will admit to find out attention to drug
discovery targeting gene expression regulation mechanism.
TF2&9
T
FI
&3
O,n,
N
<t
wo,k of
R"
~
TF I&4
TF7&8
C:V
Gene Net work
of
Human
Figure
10
Comparison gene network based on transcriptional factors
References
l.
F.
S.
Collins, E.S. Lander,
1.
Rogers, R.H. Waterston, Finishing the
euchromatic sequence
of
the human genome (2004), Nature, 431, 931-945.
2.
R.
Anat, Y. Daniei, and B. Yoav, Identifying differentially expressed genes
using false discovery rate controlling procedure,
(2003),19, Bioninformatics,
368-375.
3.
E.
Wingender, A.E. Kel,
O.
V.
Kel, H. Karas,
T.
Heinrmeyer, P. Dietze, R.
Knuppel,
A.
G.
Romaschenko and N. A. Kolchanov, TRANSFAC, TRRD
and
COMPEL: towards a federated database system on transcriptional
regulation, (1997),
Nucleic Acids Res., 25, 265-268.
4.
1.
C.
Bryne, E. Valen, M. H. Tnag, T. Marstrand, O. Winther,
1.
da Piedade,
A. Krogh,
B.
Lenhard and A. Sandelin, JASP AR, the open access database
of
transcriptional factor-binding profiles: new content and tools
in
the 2008
update, (2008), Nucleic
Acid
Res., 36,
Dl
03-1 06.
5. The FANTOM Consortium and the Riken Omics Science Center, The
transcriptional network that controls growth arrest and differentiation
in
a
human myeloid leukemia cell line,
(2009), Natur
e,
41
, 553-562
6.
S. Liang,
S.
Fuhrman, R. Somogyi, REVEAL, A GENERAL REVERSE
ENGINEERING ALGORITHM
FOR INFERENCE OF GENETIC
NETWORK ARCHITECTURES, (1998), Pacifis Symposium on
Biocomputing,
3, 18-29.
7. http://pharmacoinformatics.
jp
/cis
8. H.Sugawara,
O.
Ogasawara,
K.
Okubo,
T.
Gojobori and
Y.
Tateno: DDBJ
with new system and face, (2007), Nucleic
Acid
Res., 1-3.
9.
B.
B.
Tuch, Hli, A.D. Johnson, Evolution
of
Eukaryotic Transcription
Circuits,
(2008), SCIENCE, 319, 1797-1799

28
10.
M.
Z.
Ludwig,
C.
Bergman,
N.
H.
Patel,
M.
Kreitman,Evidence for
stabilizing selsction in a eukaryotic enhancer element
(2000), Nature,403,
564-567

Quantum
Bio-Informatics
IV
eds. L.
Accardi,
W.
Freudenb
erg
and
M.
Ohya
© 2011
World
Scientific
Publishing
Co. (pp.
29-39)
ON
LIE
GROUP-LIE
ALGEBRA
CORRESPONDENCES
OF
UNITARY
GROUPS
IN
FINITE
VON
NEUMANN
ALGEBRAS
HIROSHI
ANDO*
AND
IZUMI
OJIMAt
Research Institute for Mathematical Sciences, Graduate School
of
Science,
Kyoto
University,
Sakyo-ku, Kyoto, 606-8502, Japan
FAX
: 075-753-7272, TEL: 075-753
-7
202
* E-mail: andonuts@kurims.kyoto-u.ac.jp
t E-mail: ojima@kurims.kyoto-u.ac.jp
YASUMICHI MATSUZAWA
Departm
ent
of
Mathematics, Hokkaido University
Kita
10, Nishi 8, Kita-ku, Sapporo, 060-0810, Japan
E-mail: matsuzawa@math.sci.hokudai.ac.jp
M athematisches Institut,
Universitiit Leipzig
Johannisgasse
26, 04103, Leipzig,
Germany
This
article
is a
summary
of
our
talk
in
QBIC2010.
We give
an
affirmative
answer
to
the
question
whether
there
exist
Lie
algebras
for
suitable
closed
subgroups
of
the
unitar
y
gro
up
U(H)
in a
Hilbert
space
H
with
U(H)
equipped
with
the
stro
ng
operator
topology.
More
precisely
, for
any
stro
ngly closed
subgroup
G
of
the
unitar
y
group
U(9J1)
in a finite
von
Neumann
algebra
9J1,
we
show
that
the
set
of
all
generators
of
strongly
continuous
one-parameter
subgroups
of
G forms a
complet
e
topological
Lie
algebra
with
r
espect
to
the
stron
g
resolvent
topology.
We
also
characterize
the
algebra
9J1
of
all
densely
defined
closed
operators
affiliated
with
9J1
from
the
viewpoint
of a
tensor
category.
Keywords:
finite
von
Neumann
algebra
,
unitary
group,
affiliated
oper-
ator,
measurable
operator,
strong
resolvent
topology,
tensor
categor
y,
infinite
dimensional
Lie
group,
infinit
e
dimensional
Lie a
lgebra.
1.
Introduction
In
this
article we discuss
the
new
kind
of
Lie algebras
of
unbounded
opera-
tors. To
motivate
our
discussion, let us
start
with
the
classical
problem
of
considering
an
infinite dimensional
unitary
group
as
a Lie group.
Let
1i
be
29
