Polaina J., MacCabe A.P. (ed.). Industrial Enzymes. Structure, Function and Applications
Подождите немного. Документ загружается.

10 MACHOVI
ˇ
C AND JANE
ˇ
CEK
As regards sequence, these two types of amylase do not contain any of the
conserved regions characteristic of the -amylase family (Fig. 2). Although they are
both exo-amylases their amino acid sequences and three-dimensional structures are
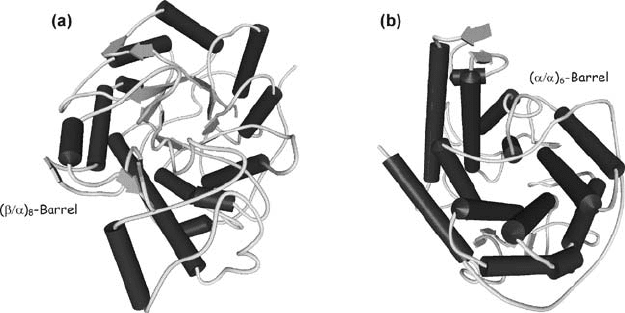
different (Aleshin et al., 1992; Mikami et al., 1993). Structurally, -amylase (Fig. 5a)
ranks along with -amylase among the large family of parallel (/
8
-barrel
proteins (Pujadas and Palau, 1999), while glucoamylase (Fig. 5b) belongs
to a smaller family of proteins adopting the (/
6
-barrel fold (Aleshin et al., 1992).
Family GH14 includes -amylases (EC 3.2.1.2) and hypothetical proteins with
sequence similarity to -amylases. Half of the family members are experimentally
verified enzymes having -amylase activity. -Amylases are especially produced
by plants: Arabidopsis thaliana, Oryza sativa, Triticum aestivum and Solanum
tuberosum. Family GH15 includes glucoamylases (EC 3.2.1.3), two glucodex-
tranases (EC 3.2.1.70) and hypothetical proteins with sequence similarity to GH15.
Again, about 50% of the family members are experimentally verified enzymes
having glucoamylase or glucodextranase activities.
The first determined three-dimensional structure of a -amylase was that of
soybean (Mikami et al., 1993). At present, the structures of -amylases from
sweet potato (Cheong et al., 1995), barley (Mikami et al., 1999b) and Bacillus
cereus (Mikami et al., 1999a; Oyama et al., 1999) are also known. The core of
the -amylase structure is formed by the catalytic (/
8
-barrel domain (Fig. 5a)
followed by the C-terminal loop region. Although this loop surrounds the N-terminal
side of the (/
8
-barrel and may stabilise the whole -amylase molecule, it is
not involved in catalysis (Mikami, 2000). As has been pointed out above, the
-amylase (/
8
-barrel differs from that of -amylase and all other enzymes
of clan GH-H, resembling more the single-domain structure of triosephosphate
isomerase (Mikami, 2000). The two amino acid residues responsible for catalysis are
the two glutamates, Glu186 and Glu380 (soybean -amylase numbering), positioned
Figure 5. Three-dimensional structures of (a) GH14 -amylase from soybean (1BYA; Mikami
et al., 1993) and (b) GH15 glucoamylase from Aspergillus awamori (1AGM; Aleshin et al., 1992)
AMYLOLYTIC ENZYMES 11
near the C-terminus of strands 4 and 7 of the (/
8
-barrel domain, respec-
tively (Mikami et al., 1994). Totsuka and Fukazawa (1996) described further the
indispensable roles for Asp101 and Leu383 in addition to the two catalytic gluta-
mates. Analyses of the (/
8
-barrel fold of -amylases from both the evolutionary
and structural points of view are available (Pujadas et al., 1996; Pujadas and
Palau, 1997).
Glucoamylase structures have been solved for two fungal enzymes: Aspergillus
awamori (Aleshin et al., 1992) and the yeast Saccharomycopsis fibuligera (Sevcik
et al., 1998), and one bacterial enzyme from Thermoanaerobacterium thermosac-
charolyticum (Aleshin et al., 2003). The glucoamylase catalytic domain is composed
of 12 -helices that form the so-called (/
6
-barrel fold (Fig. 5b). It consists of
an inner core of six mutually parallel -helices that are connected to each other
through a peripheral set of six -helices which are parallel to each other but approx-
imately antiparallel to the inner core of the -helices (Aleshin et al., 1992). This
fold is not as frequent as the TIM-barrel fold (Farber and Petsko, 1990; Jane
ˇ
cek
and Bateman, 1996; Pujadas and Palau, 1999), however, the (/
6
-barrel has also
been found in different proteins and enzymes, for example in the enzymes from
families GH8 and GH9 (Juy et al., 1992; Alzari et al., 1996). Some glucoamylases,
like some -amylases (and related enzymes from the clan GH-H) and -amylases,
contain starch-binding domains (Svensson et al., 1989; Jane
ˇ
cek and Sevcik, 1999)
which can be of various types (for a review, see Rodriguez-Sanoja et al., 2005).
The starch-binding domain may be evolutionarily independent from the catalytic
domain (Jane
ˇ
cek et al., 2003). It should also be possible to add a starch-binding
domain artificially to an amylase (or eventually to any other protein) to improve its
amylolytic and raw starch-binding and degradation abilities (Ohdan et al., 2000; Ji
et al., 2003; Hua et al., 2004; Levy et al., 2004; Kramhøft et al., 2005; Latorre-
Garcia et al., 2005). Recently, it seems evident that some amylases may contain
starch-binding activity without a specific structural module (Hostinova et al., 2003;
Tranier et al., 2005).
Based on the analysis of glucoamylase amino acid sequences, Coutinho and
Reilly (1997) described seven subfamilies taxonomically corresponding to bacterial
(1), archaeal (1), yeast (3) and fungal (2) origins. As evidenced by the crystal
structures of the glucoamylases from Aspergillus awamori (Harris et al., 1993;
Aleshin et al., 1994, 1996; Stoffer et al., 1995) and Saccharomycopsis fibuligera
(Sevcik et al., 1998), the two glutamates, Glu179 and Glu400 (Aspergillus
enzyme numbering), act as the key catalytic residues. The next most well-studied
glucoamylase is that from Aspergillus niger (Christensen et al., 1996; Frandsen
et al., 1996) which is highly similar to the Aspergillus awamori counterpart.
5. FAMILY GH31
There are some glucoamylases that have been classified into family GH31 together
with -glucosidases, -xylosidases and glucan lyases (Yu et al., 1999; Lee
et al., 2003; 2005b). These enzymes act through a retaining mechanism like the
12 MACHOVI
ˇ
C AND JANE
ˇ
CEK
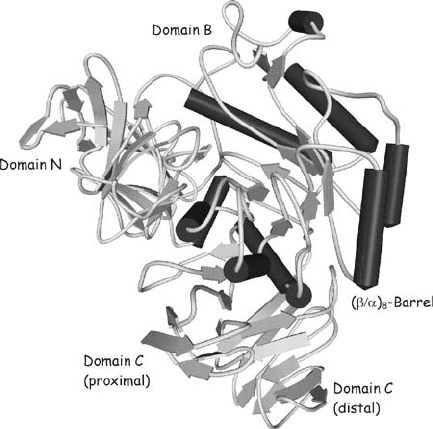
Figure 6. Three-dimensional structure of GH31 -xylosidase from Escherichia coli (1XSI;
Lovering et al., 2005)
members of clan GH-H (Chiba , 1997; Nakai et al., 2005). GH31 was considered to
be a member of clan GH-H because of remote sequence homologies between GH31
and GH13 enzymes (Rigden, 2002). This assumption has recently been supported
by the resolution of the three-dimensional structure of a GH31 -xylosidase from
Escherichia coli (Lovering et al., 2005) and -glucosidase from Sulfolobus solfa-
taricus (Ernst et al., 2006) showing the expected (/
8
-barrel catalytic domain
(Fig. 6). Interestingly, the domain arrangement of the GH31 members strongly
resembles that of GH13 enzymes (Fig. 3), especially regarding domain B protruding
out of the (/
8
-barrel in the place of loop 3 (Lovering et al., 2005).
6. FAMILY GH57
For a long time GH57 has been one of the most popular GH families, attracting
much scientific interest. More than 15 years ago the sequence of a heat-stable
-amylase from the thermophilic bacterium Dictyoglomus thermophilum was
published (Fukusumi et al., 1988). Despite the fact that this sequence encoded an -
amylase, its analysis did not reveal any detectable similarity with GH13 -amylases.
Later, a similar sequence encoding the -amylase from the hyperthermophilic
archaeon, Pyrococcus furiosus, was determined (Laderman et al., 1993). These two
sequences became the basis for the new amylolytic family, GH57, established in
1996 (Henrissat and Bairoch 1996). In the last few years, when entire genomes
AMYLOLYTIC ENZYMES 13
of many micro-organisms have been sequenced, family GH57 has expanded. Its
members are all prokaryotic enzymes, most of them from hyperthermophilic archaea
(Zona et al., 2004). At present the GH57 family consists of about 100 members
(Coutinho and Henrissat, 1999) and five enzyme (Jane
ˇ
cek, 2005; Murakami et al.,
2006): -amylase (EC 3.2.1.1), -galactosidase (EC 3.2.1.22), amylopullulanase
(EC 3.2.1.1/41), branching enzyme (EC 2.4.1.18) and 4--glucanotransferase (EC
2.4.1.25). Only about 10% of the family sequence entries are enzymes; all others are
hypothetical proteins without known activity (Zona et al., 2004). GH57 sequences
are highly heterogeneous: some of them have less than 400 residues whereas others
have more than 1,500 residues (Zona et al., 2004).
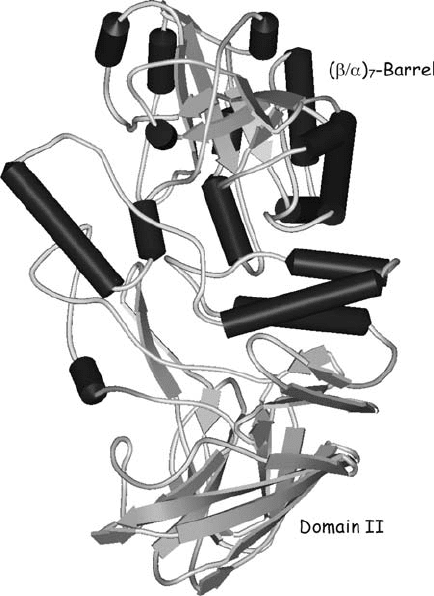
Structural information for GH57 members is scarce. To date, only the structures
of the 4--glucanotransferase from Thermococcus litoralis (Imamura et al., 2003)
and AmyC enzyme from Thermotoga maritima (Dickmanns et al., 2006) have been
determined. They both revealed a (/
7
-barrel fold (Fig. 7), i.e. an incomplete
TIM-barrel. Glu123 and Asp214 (T. litoralis enzyme numbering) which define the
Figure 7. Three-dimensional structure of GH57 4--glucanotransferance from Thermococcus litoralis
(1K1W; Imamura et al., 2003)
14 MACHOVI
ˇ
C AND JANE
ˇ
CEK
catalytic centre of the enzyme, are arranged at a distance of less than 7 Å (Imamura
et al., 2003), thus confirming that GH57 also employs a retaining mechanism for
-glycosidic bond cleavage.
New information about GH57 has arisen from a bioinformatic study focused on
the conserved sequences containing the pair of catalytic residues (Zona et al., 2004).
In addition to T. litoralis 4--glucanotransferase, both catalytic residues were
experimentally identified in two amylopullulanases from Thermococcus hydrother-
malis (Zona et al., 2004) and Pyrococcus furiosus (Kang et al., 2005). The catalytic
nucleophile was found also in the -galactosidase from Pyrococcus furiosus (Van
Lieshout et al., 2003). Biochemical analysis indicates that family GH57 enzymes
may lack a genuine -amylase specificity (Jane
ˇ
cek, 2005).
REFERENCES
Aghajari, N., Feller, G., Gerday, C., and Haser, R. (1998). Structures of the psychrophilic Alteromonas
haloplanctis -amylase give insights into cold adaptation at a molecular level. Structure 6, 1503–1516.
Aleshin, A.E., Feng, P.H., Honzatko, R.B., and Reilly, P.J. (2003). Crystal structure and evolution of
prokaryotic glucoamylase. J. Mol. Biol. 327, 61–73.
Aleshin, A.E., Firsov, L.M., and Honzatko, R.B. (1994). Refined structure for the complex of acarbose
with glucoamylase from Aspergillus awamori var. X100 to 2.4 Å resolution. J. Biol. Chem. 269,
15631–15639.
Aleshin, A.E., Golubev, A., Firsov, L.M., and Honzatko, R.B. (1992). Crystal structure of glucoamylase
from Aspergillus awamori var. X100 to 2.2 Å resolution. J. Biol. Chem. 267, 19291–19298.
Aleshin, A.E., Stoffer, B., Firsov, L.M., Svensson, B., and Honzatko, R.B. (1996). Crystallographic
complexes of glucoamylase with maltooligosaccharide analogs: relationships of stereochemical distor-
sions at the nonreducing end to the catalytic mechanism. Biochemistry 35, 8319–8328.
Alzari, P.M., Souchon, H., and Dominguez, R. (1996). The crystal structure of endoglucanase CelA,
a family 8 glycosyl hydrolase from Clostridium thermocellum. Structure 4, 265–275.
Argüello-Morales, M.A., Renaud-Simeon, M., Pizzut, S., Sarcabal, P., Willemot, R.M., and Monsan, P.
(2000). Sequence analysis of the gene encoding alternansucrase from Leuconostoc mesenteroides
NRRLB-1355. FEMS Microbiol. Lett. 182, 81–85.
Ball, S., Guan, H.P., James, M., Myers, A., Keeling, P., Mouille, G., Buléon, A., Colonna, P., and
Preiss, J. (1996). From glycogen to amylopectin: a model for the biogenesis of the plant starch granule.
Cell 86, 349–352.
Banner, D.W., Bloomer, A., Petsko, G.A., Phillips, D.C., and Wilson, I.A. (1975). Atomic coordinates
for triose phosphate isomerase from chicken muscle. Biochem. Biophys. Res. Commun. 72, 146–155.
Buisson, G., Duee, E., Haser, R., and Payan, F. (1987). Three dimensional structure of porcine pancreatic
-amylase at 2.9 Å resolution. Role of calcium in structure and activity. EMBO J. 6, 3909–3916.
Cheong, C.G., Eom, S.H., Chang, C., Shin, D.H., Song, H.K., Min, K., Moon, J.H., Kim, K.K., Hwang,
K.Y., and Suh, S. W. (1995). Crystallization, molecular replacement solution, and refinement of
tetrameric -amylase from sweet potato. Proteins 21, 105–117.
Chiba, S. (1997). Molecular mechanism in -glucosidase and glucoamylase. Biosci. Biotechnol.
Biochem. 61, 1233–1239.
Christensen, U., Olsen, K., Stoffer, B.B., and Svensson, B. (1996). Substrate binding mechanism of
Glu180 → Gln, Asp176 → Asn, and wild-type glucoamylases from Aspergillus niger. Biochemistry
35, 15009–15018.
Coutinho, P.M., and Henrissat, B. (1999). Glycoside hydrolase family server. URL: http:
//afmb.cnrs-mrs.fr/_pedro/CAZY/ghf.html
Coutinho, P.M., and Reilly, P.J. (1997). Glucoamylase structural, functional, and evolutionary relation-
ships. Proteins 29, 334–347.
AMYLOLYTIC ENZYMES 15
Davies, G., and Henrissat, B. (1995). Structures and mechanisms of glycosyl hydrolases. Structure 3,
853–859.
Devulapalle, K.S., Goodman, S.D., Gao, Q., Emsley, A. and Mooser, G. (1997). Knowledge-based
model of a glucosyltransferase from the oral acterial group of mutans streptococci. Protein Sci. 6,
2489–2493.
Dickmanns, A., Ballschmiter, M., Liebl, W., and Ficner, R. (2006). Structure of the novel -amylase
AmyC from Thermotoga maritima. Acta Crystallogr. D Biol. Crystallogr. 62, 262–270.
Egloff, M.P., Uppenberg, J., Haalck, L., and Van Tilbeurgh, H. (2001). Crystal structure of maltose
phosphorylase from Lactobacillus brevis: unexpected evolutionary relationship with glucoamylases.
Structure 9, 689–697.
Ernst, H.A., Lo Leggio, L., Willemoes, M., Leonard, G., Blum, P., and Larsen, S. (2006). Structure of the
Sulfolobus solfataricus -glucosidase: implications for domain conservation and substrate recognition
in GH31. J. Mol. Biol. 358, 1106–1124.
Farber, G.K., and Petsko G.A. (1990). The evolution of / barrel enzymes. Trends Biochem. Sci. 15,
228–234.
Frandsen, T.P., Stoffer, B.B., Palcic, M.M., Hof, S., and Svensson, B. (1996). Structure and energetics
of the glucoamylase-isomaltose transition-state complex probed by using modeling and deoxygenated
substrates coupled with site-directed mutagenesis. J. Mol. Biol. 263, 79–89.
Fraser, C.M., Casjens, S., and Venter, J.C. (1997). Genomic sequence of a Lyme disease spirochaete,
Borrelia burgdorferi. Nature 390, 580–586.
Fukusumi, S., Kamizono, A., Horinouchi, S., and Beppu, T. (1988). Cloning and nucleotide sequence
of a heat-stable amylase gene from an anaerobic thermophile, Dictyoglomus thermophilum. Eur. J.
Biochem. 174, 15–21.
Glöckner, G., Lehmann, R., Romualdi, A., Pradella, S., Schulte-Spechtel, U., Schilhabel, M., Wilske,
B. Sühnel, J., and M. Platzer (2004). Comparative analysis of the Borrelia garinii genome. Nucleic
Acids Res. 32, 6038–6046.
Godany, A., Vidova, B., Bhide, M., Tkacikova, L., and Jane
ˇ
cek, S. (2005). A unique member of the
-amylase family, the amylomaltase-like protein (BB0166) from Borrelia burgdorferi homologous to
Thermus aquaticus amylomaltase, contains conserved only the catalytic triad. In: Thermophiles 05,
International Conference, September 18-22, Gold Coast, Australia, pp. 149–150.
Harris, E. M. S., Aleshin, A.E., Firsov, L.M., and Honzatko, R.B. (1993). Refined structure for the
complex of 1-deoxynojirimycin with glucoamylase from Aspergillus awamori var. X100 to 2.4
Å resolution. Biochemistry 32, 1618–1626.
Henrissat, B. (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities.
Biochem. J. 280, 309–316.
Henrissat, B., and Bairoch, A. (1996). Updating the sequence-based classification of glycosyl hydrolases.
Biochem. J. 316, 695–696.
Hostinova, E., Solovicova, A., Dvorsky, R., and Gasperik, J. (2003). Molecular cloning and 3D structure
prediction of the first raw-starch-degrading glucoamylase without a separate starch-binding domain.
Arch. Biochem. Biophys. 411, 189–195.
Hua, Y.W., Chi, M.C., Lo, H.F., Hsu, W.H., and Lin, L.L. (2004). Fusion of Bacillus stearothermophilus
leucine aminopeptidase II with the raw-starch-binding domain of Bacillus sp. strain TS-23 -amylase
generates a chimeric enzyme with enhanced thermostability and catalytic activity. J. Ind. Microbiol.
Biotechnol. 31, 273–277.
Imamura, H., Fushinobu, S., Yamamoto, M., Kumasaka, T., Jeon, B.S., Wakagi, T., and Matsuzawa,
H. (2003). Crystal structures of 4--glucanotransferase from Thermococcus litoralis and its complex
with an inhibitor. J. Biol. Chem. 278, 19378–19386.
Jane
ˇ
cek, S. (1994). Parallel /-barrels of -amylase, cyclodextrin glycosyltransferase and oligo-1,6-
glucosidase versus the barrel of -amylase: evolutionary distance is a reflection of unrelated sequences.
FEBS Lett. 353, 119–123.
Jane
ˇ
cek, S. (1997). -Amylase family: molecular biology and evolution. Progr. Biophys. Mol. Biol. 67,
67–97.
16 MACHOVI
ˇ
C AND JANE
ˇ
CEK
Jane
ˇ
cek, S. (2000). How many conserved sequence regions are there in the -amylase family? Biologia,
Bratislava 57 (Suppl. 11), 29–41.
Jane
ˇ
cek, S. (2005). Amylolytic families of glycoside hydrolases: focus on the family GH-57. Biologia,
Bratislava 60 (Suppl. 16), 177–184.
Jane
ˇ
cek, S., and Bateman, A. (1996). The parallel (/
8
-barrel: perhaps the most universal and the
most puzzling protein folding motif. Biologia. Bratislava 51, 613–628.
Jane
ˇ
cek, S., and Sevcik, J. (1999). The evolution of starch-binding domain. FEBS Lett. 456, 119–125.
Jane
ˇ
cek, S., Svensson, B., and MacGregor, E. A. (2003). Relation between domain evolution, specificity,
and taxonomy of the -amylase family members containing a C-terminal starch-binding domain. Eur.
J. Biochem. 270, 635–645.
Jespersen, H., MacGregor, E.A., Sierks, M.R., and Svensson, B. (1991). Comparison of the domain-level
organization of starch hydrolases and related enzymes. Biochem. J. 280, 51–55.
Ji, Q., Vincken, J.P., Suurs, L.C., and Visser, R.G. (2003). Microbial starch-binding domains as a tool
for targeting proteins to granules during starch biosynthesis. Plant Mol. Biol. 51, 789–801.
Juy, M., Amit, A.G., Alzari, P.M., Poljak, R.J., Clayssens, M., Béguin, P., and Aubert J.P. (1992).
Three-dimensional structure of a thermostable bacterial cellulase. Nature 357, 89–91.
Kang, S., Vieille, C., and Zeikus, J.G. (2005). Identification of Pyrococcus furiosus amylopullulanase
catalytic residues. Appl. Microbiol. Biotechnol. 66, 408–413.
Kramhøft, B., Bak-Jensen, K.S., Mori, H., Juge, N., Nøhr, J., and Svensson, B. (2005). Involvement
of individual subsites and secondary substrate binding sites in multiple attack on amylose by barley
-amylase. Biochemistry 44, 1824–1832.
Kuriki, T. (2000). Catalytic mechanism of glycoenzymes. In: Glycoenzymes (Eds. M. Ohnishi,
T. Hayashi, S. Ishijima, T. Kuriki), pp. 3–17, Japan Scientific Societies Press, Tokyo.
Kuriki, T., and Imanaka, T. (1999). The concept of the -amylase family: structural similarity and
common catalytic mechanism. J. Biosci. Bioeng. 87, 557–565.
Laderman, K.A., Asada, K., Uemori, T., Mukai, H., Taguchi, Y., Kato, I., and Anfinsen, C.B. (1993). -
amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus – cloning and sequencing
of the gene and expression in Escherichia coli. J. Biol. Chem. 268, 24402–24407.
Latorre-Garcia, L., Adam, A.C., Manzanares, P., and Polaina, J. (2005). Improving the amylolytic activity
of Saccharomyces cerevisiae glucoamylase by the addition of a starch binding domain. J. Biotechnol.
118, 167–176.
Lee, H.S., Kim, J.S., Shim, K.H., Kim, J.W., Park, C.S., and Park, K.H. (2005a). Quaternary structure and
enzymatic properties of cyclomaltodextrinase from alkalophilic Bacillus sp. I-5. Biologia, Bratislava
60 (Suppl. 16), 73–77.
Lee, S.S., Yu, S., and Withers, S.G. (2003). Detailed dissection of a new mechanism for glycoside
cleavage: -1,4-glucan lyase. Biochemistry 42, 13081–13090.
Lee, S.S., Yu, S., and Withers, S. G. (2005b). Mechanism of action of exo-acting -1,4-glucan lyase: a
glycoside hydrolase family 31 enzyme. Biologia Bratislava 60 (Suppl. 16), 137–148.
Levy, I., Paldi, T., and Shoseyov, O. (2004). Engineering a bifunctional starch-cellulose cross-bridge
protein. Biomaterials 25, 1841–1849.
Libessart, N., and Preiss, J. (1998). Arginine residue 384 at the catalytic center is important for branching
enzyme II from maize endosperm. Arch. Biochem. Biophys. 360, 135–141.
Lovering, A.L., Lee, S.S., Kim, Y.W., Withers, S.G., and Strynadka, N. C. J. (2005). Mechanistic and
structural analysis of a family 31 -glycosidase and its glycosyl-enzyme intermediate. J. Biol. Chem.
280, 2105–2115.
MacGregor, E. A. (2005). An overview of clan GH-H and distantly-related families. Biologia Bratislava
60 (Suppl. 16), 5–12.
MacGregor, E.A., Jane
ˇ
cek, S., and Svensson, B. (2001). Relationship of sequence and structure to
specificity in the -amylase family of enzymes. Biochim. Biophys. Acta 1546, 1–20.
MacGregor, E.A., Jespersen, H.M., and Svensson, B. (1996). A circularly permuted -amylase-type
/-barrel structure in glucan-synthesizing glucosyltransferases. FEBS Lett. 378, 263–266.
Machius, M., Wiegand, G., and Huber, R. (1995). Crystal structure of calcium-depleted Bacillus licheni-
formis -amylase at 2.2 Å resolution. J. Mol. Biol. 246, 545–559.
AMYLOLYTIC ENZYMES 17
Machovi
ˇ
c, M., and Jane
ˇ
cek, S. (2003). The invariant residues in the -amylase family: just the catalytic
triad. Biologia, Bratislava 58, 1127–1132.
Matsuura, Y. (2002). A possible mechanism of catalysis involving three essential residues in the enzymes
of -amylase family. Biologia Bratislava, 57 (Suppl. 11), 21–27.
Matsuura, Y., Kusunoki, M., Harada, W., and Kakudo, M. (1984). Structure and possible catalytic
residues of Taka-amylase A. J. Biochem. Tokyo 95, 697–702.
Mikami, B. (2000). Structure of -amylase: X-ray crystallographic analysis. In: Glycoenzymes (Eds.
M. Ohnishi, T. Hayashi, S. Ishijima, T. Kuriki), 55–81, Japan Scientific Societies Press, Tokyo
Mikami, B., Adachi, M., Kage, T., Sarikaya, E., Nanmori, T., Shinke, R., and Utsumi, S. (1999a).
Structure of raw starch-digesting Bacillus cereus -amylase complexed with maltose. Biochemistry
38, 7050–7061.
Mikami, B., Degano, M., Hehre, E.J., and Sacchettini, J.C. (1994). Crystal structures of soybean
-amylase reacted with -maltose and maltal: active site components and their apparent roles in
catalysis. Biochemistry 33, 7779–7787.
Mikami, B., Hehre, E.J., Sato, M., Katsube, Y., Hirose, M., Morita, Y., and Sacchettini, J. C. (1993).
The 2.0 Å resolution structure of soybean -amylase complexed with -cyclodextrin. Biochemistry
32, 6836–6845.
Mikami, B., Yoon, H.J., and Yoshigi, N. (1999b). The crystal structure of the sevenfold mutant of barley
-amylase with increased thermostability at 2.5 Å resolution. J. Mol. Biol. 285, 1235–1243.
Mouille, G., Maddelein, M.L., Libessart, N., Talaga, P., Decq, A., Delrue, B., and Ball, S. (1996).
Preamylopectin processing: a mandatory step for starch biosynthesis in plants. Plant Cell 8, 1353–1366.
Murakami, T., Kanai, T., Takata, H., Kuriki, T., and Imanaka, T. (2006). A novel branching enzyme
of the GH-57 family in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J.
Bacteriol. 188, 5915–5924.
Nakai, H., Okuyama, M., Kim, Y.M., Saburi, W., Wongchawalit, J., Mori, H., Chiba, S., and Kimura, A.
(2005). Molecular analysis of -glucosidase belonging to GH-family 31. Biologia Bratislava 60
(Suppl. 16), 131–135.
Nakajima, R., Imanaka, T., and Aiba, S. (1986). Comparison of amino acid sequences of eleven different
-amylases. Appl. Microbiol. Biotechnol. 23, 355–360.
Oh, B.H. (2003). The same group of enzymes with different names: cyclomaltodextrinases, neopullu-
lanases, and maltogenic amylases. Biologia, Bratislava 58, 299–305.
Ohdan, K., Kuriki, T., Takata, H., Kaneko, H., and Okada, S. (2000). Introduction of raw starch-
binding domains into Bacillus subtilis -amylase by fusion with the starch-binding domain of Bacillus
cyclomaltodextrin glucanotransferase. Appl. Environ. Microbiol. 66, 3058–3064.
Oslancova, A., and Jane
ˇ
cek, S. (2002). Oligo-1,6-glucosidase and neopullulanase enzyme subfamilies
from the -amylase family defined by the fifth conserved sequence region. Cell. Mol. Life Sci. 59,
1945–1959.
Oyama, T., Kusunoki, M., Kishimoto, Y., Takasaki, Y., and Nitta, Y. (1999). Crystal structure of -amylase
from
Bacillus cereus var. mycoides at 2.2 Å resolution. J. Biochem. (Tokyo) 125, 1120–1130.
Przylas, I., Tomoo, K., Terada, Y., Takaha, T., Fujii, K., Saenger, W., and Sträter, N. (2000). Crystal
structure of amylomaltase from Thermus aquaticus, a glycosyltransferase catalysing the production
of large cyclic glucans. J. Mol. Biol. 296, 873–886.
Pujadas, G., and Palau, J. (1997). Anatomy of a conformational transition of -strand 6 in soybean
-amylase caused by substrate (or inhibitor) binding to the catalytical site. Protein Sci. 6, 2409–2417.
Pujadas, G., and Palau, J. (1999). TIM barrel fold: structural, functional and evolutionary characteristics
in natural and designed molecules. Biologia, Bratislava 54, 231–254.
Pujadas, G., and Palau, J. (2001). Evolution of -amylases: architectural features and key residues in
the stabilization of the (/
8
scaffold. Mol. Biol. Evol. 18, 38–54.
Pujadas, G., Ramirez, F. M., Valero, R., Palau, J. (1996). Evolution of -amylase: patterns of variation
and conservation in subfamily sequences in relation to parsimony mechanisms. Proteins 25, 456–472.
Reardon, D., and Farber, G.K. (1995). The structure and evolution of / barrel proteins. FASEB J. 9,
497–503.
18 MACHOVI
ˇ
C AND JANE
ˇ
CEK
Rigden, D.J. (2002). Iterative database searches demonstrate that glycoside hydrolase families 27, 31,
36 and 66 share a common evolutionary origin with family 13. FEBS Lett. 53, 17–22.
Robyt, J. F. (2005). Inhibition, activation, and stabilization of -amylase family enzymes. Biologia,
Bratislava 60 (Suppl. 16), 17–26.
Rodriguez-Sanoja, R., Oviedo, N., and Sanchez, S. (2005). Microbial starch-binding domain. Curr. Opin.
Microbiol. 8, 260–267.
Sinnot, M.L. (1990). Catalytic mechanisms of enzymatic glycosyl transfer. Chem. Rev. 90, 1171–1202.
Søgaard, M., Kadziola, A., Haser, R., and Svensson, B. (1993). Site-directed mutagenesis of histidine
93, aspartic acid 180, glutamic acid 205, histidine 290, and aspartic acid 291 at the active site and
tryptophan 279 at the raw starch binding site in barley -amylase 1. J. Biol. Chem. 268, 22480–22484.
Stoffer, B., Aleshin, A.E., Firsov, L.M., Svensson, B., and Honzatko, R. B. (1995). Refined structure for
the complex of D-gluco-dihydroacarbose with glucoamylase from Aspergillus awamori var. X100 to
2.2 Å resolution: dual conformations for extended inhibitors bound to the active site of glucoamylase.
FEBS Lett. 358, 57–61.
Svensson, B. (1994). Protein engineering in the -amylase family: catalytic mechanism, substrate
specificity, and stability. Plant Mol. Biol. 25, 141–157.
Svensson, B., Jensen, M.T., Mori, H., Bak-Jensen, K.S., Bønsager, B., Nielsen, P.K., Kramhøft, B.,
Prætorius-Ibba, M., Nøhr, J., Juge, N., Greffe, L., Williamson, G., and Driguez, H. (2002). Facinating
facets of function and structure of amylolytic enzymes of glycoside hydrolase family 13. Biologia,
Bratislava 57 (Suppl. 11), 5–19.
Svensson, B., Jespersen, H., Sierks, M.R., and MacGregor, E.A. (1989). Sequence homology between
putative raw starch-binding domains from different starch degrading enzymes. Biochem. J. 264,
309–311.
Sevcik, J., Solovicova, A., Hostinova, E., Gasperik, J., Wilson, K.S., and Dauter, Z. (1998). Structure
of glucoamylase from Saccharomycopsis fibuligera at 1.7 Å resolution. Acta Crystallogr. D Biol.
Crystallogr. 54, 854–866.
Totsuka, A., and Fukazawa, C. (1996). Functional analysis of Glu380 and Leu383 of soybean -amylase.
A proposed action mechanism. Eur. J. Biochem. 240, 655–659.
Tranier, S., Deville, K., Robert, X., Bozonnet, S., Haser, R., Svensson, B., and Aghajari, N. (2005).
Insights into the “pair of sugar tongs” surface binding site in barley -amylase isozymes and crystal-
lization of appropriate sugar tongs mutants. Biologia, Bratislava 60 (Suppl. 16), 37–46.
Turner, P., Nilsson, C., Svensson, D., Holst, O., Gorton, L., and Nordberg Karlsson, E. (2005).
Monomeric and dimeric cyclomaltodextrinases reveal different modes of substrate degradation.
Biologia, Bratislava 60 (Suppl. 16), 79–87.
Van Lieshout, J.F.T., Verhees, C.H., Ettema, T.J.G., van der Saar, S., Imamura, H., Matsuzawa, H., van
der Oost, J., and de Vos, W.M. (2003). Identification and molecular characterization of a novel type
of -galactosidase from Pyrococcus furiosus. Biocatal. Biotransform. 21, 243–252.
Vihinen, M., and Mäntsala, P. (1989). Microbial amylolytic enzymes. Crit. Rev. Biochem. Mol. Biol.
24, 329–418.
Vihinen, M., Ollikka, P., Niskanen, J., Meyer, P., Suominen, I., Karp, M., Holm, L., Knowles, J., and
Mäntsälä, P. (1990). Site-directed mutagenesis of a thermostable -amylase from Bacillus stearother-
mophilus: putative role of three conserved residues. J. Biochem. 107, 267–272.
Yu, S., Bojsen, K., Svensson, B., and Marcussen, J. (1999). -1,4-glucan lyases producing 1,5-anhydro-
D-fructose from starch and glycogen have sequence similarity to -glucosidases. Biochim. Biophys.
Acta
1433, 1–15.
Zona, R., Chang-Pi-Hin, F., O’Donohue, M.J., and Jane
ˇ
cek, S. (2004). Bioinformatics of the glycoside
hydrolase family 57 and identification of catalytic residues in amylopullulanase from Thermococcus
hydrothermalis. Eur. J. Biochem. 271, 2863–2872.
CHAPTER 2
THE USE OF STARCH PROCESSING ENZYMES
IN THE FOOD INDUSTRY
JÓZEF SYNOWIECKI
∗
Department of Food Chemistry, Technology and Biotechnology, Chemical Faculty, Gdansk University
of Technology, Gdansk, Poland
∗
synowiec@chem.pg.gda.pl
1. INTRODUCTION
Starch, the main component of many agricultural products, e.g. corn (maize),
potatoes, rice and wheat, is deposited in plant cells as reserve material for the
organism in the form of granules which are insoluble in cold water. This carbo-
hydrate is the main constituent of food products such as bread and other bakery
goods or is added to many foods for its functionality as a thickener, water binder,
emulsion stabilizer, gelling agent and fat substitute. Starch granules consist of two
types of molecules composed of -D-glucose units called amylose and amylopectin.
In amylose almost all the glucose residues are linked by -1,4-glycosidic bonds,
whereas in amylopectin about 5 % of the carbohydrate units are also joined by -1,6-
linkages forming branch points. The relative contents of amylose and amylopectin
depend on the plant species. For example, wheat starch contains about 25% amylose
while waxy corn starch is more than 97–99% amylopectin. Starch origin also makes
differences to the size, shape and structure of the polysaccharide granules, their
swelling power, gelatinisation temperature, extent of esterification with phosphoric
acid, and the amounts of lipids and other compounds which are retained inside the
hydrophobic inner surface of the amylose helices.
Expanding starch functionality can be achieved through chemical or enzymatic
modifications. The most important methods of enzymatic starch processing (Fig. 1)
are the production of cyclodextrins and the hydrolysis of starch into a mixture of
simpler carbohydrates for the production of syrups having different compositions
and properties. These products are used in a wide variety of foodstuffs: soft drinks,
confectionery, meats, packed products, ice cream, sauces, baby food, canned fruit,
19
J. Polaina and A.P. MacCabe (eds.), Industrial Enzymes, 19–34.
© 2007 Springer.
