Nielsen H. RNA: Methods and Protocols
Подождите немного. Документ загружается.


244 Tenenbaum, Christiansen, and Nielsen
binding protein (SLBP) to a conserved hairpin in the 3
UTR of
the mRNAs. RIP-Chip analysis of SLBP targets identified approx-
imately 30 such mRNAs that might function as an RNA operon
for coordination of histone production during replication (30).
A broader example in the same vein is the regulation of mRNAs
that exhibit coordinated decay by the presence of AU-rich ele-
ments (AREs) in their 3
UTRs. These elements of 50–150 nt
are found in 5–8% of all mRNAs and are typically associated with
mRNAs with a short half-life that has to be tightly regulated as
part of a transient cellular response (31). AREs are bound by
different kinds of ARE-binding proteins. As an example, it has
been shown that distinct classes of mRNAs that encode regula-
tors of the immune response, such as cytokines and chemokines,
have similar half-lives that can be altered in a concerted man-
ner. RIP-Chip and other types of analyses have shown that this
can be mediated by the binding of ARE-binding proteins such
as ELAV/Hu (1) and tristetraproline (TTP). RNA operons that
are involved in decay seem to be relatively common and examples
include the decay of mRNAs involved in ribosome biogenesis and
iron metabolism (20).
4. Conclusions
The post-transcriptional operon model appears to be valuable
at our current conceptual understanding of molecular biology.
Importantly, it is associated with specific methods that provide
a new type of information that can be used to reveal unex-
pected participants in cellular pathways and deepen our under-
standing of cellular responses. Furthermore, r ecent advances in
high-throughput sequencing will make these methods even more
attractive and contribute to the generation of a knowledge plat-
form on post-transcriptional regulation that will be helpful in the
discipline of systems biology.
References
1. Tenenbaum, S. A., Carson, C. C., Lager, P.
J., Keene, J. D. (2000) Identifying mRNA
subsets in messenger ribonucleoprotein com-
plexes by using cDNA arrays. Proc Natl Acad
Sci USA 97, 14085–14090.
2. Keene, J. D. (2001) Ribonucleoprotein
infrastructure regulating the flow of genetic
information between the genome and the
proteome. Proc Natl Acad Sci USA 98,
7018–7024.
3. Keene, J. D., Tenenbaum, S. A. (2002)
Eukaryotic mRNPs may represent post-
transcriptional operons. Mol Cell 9,
1161–1167.
4. Keene, J. D. (2007) RNA regulons: coor-
dination of post-transcriptional events. Nat
Rev Genet 8, 533–543.
5. Mansfield, K. D., Keene, J. D. (2009)
The ribonome: a dominant force in co-
ordinating gene expression. Biol Cell 101,
169–181.
6. Neil, H., Malabat, C., ubenton-Carafa, Y.,
Xu, Z., Steinmetz, L. M., Jacquier, A. (2009)
Widespread bidirectional promoters are the
The Post-transcriptional Operon 245
major source of cryptic transcripts in yeast.
Nature 457, 1038–1042.
7. He, Y., Vogelstein, B., Velculescu, V. E.,
Papadopoulos, N., Kinzler, K. W. (2008) The
antisense transcriptomes of human cells. Sci-
ence 322, 1855–1857.
8. Oliver, B., Parisi, M., Clark, D. (2002) Gene
expression neighborhoods. JBiol1, 4.
9. Carninci, P. (2009) Molecular biology: the
long and short of RNAs. Nature 457,
974–975.
10. Seila, A. C., Calabrese, J. M., Levine, S. S.,
Yeo, G. W., Rahl, P. B., Flynn, R. A., Young,
R. A., Sharp, P. A. (2008) Divergent tran-
scription from active promoters. Science 322,
1849–1851.
11. Maniatis, T., Reed, R. (2002) An extensive
network of coupling among gene expression
machines. Nature 416, 499–506.
12. Moore, M. J., Proudfoot, N. J. (2009) Pre-
mRNA processing reaches back to transcrip-
tion and ahead to translation. Cell 136,
688–700.
13. Gerstein, M. B., Bruce, C., Rozowsky, J. S.,
Zheng, D., Du, J., Korbel, J. O., Emanuels-
son, O., Zhang, Z. D., Weissman, S., Snyder,
M. (2007) What is a gene, post-ENCODE?
History and updated definition. Genome Res
17, 669–681.
14. Pan, Q., Shai, O., Lee, L. J., Frey, B. J.,
Blencowe, B. J. (2008) Deep surveying of
alternative splicing complexity in the human
transcriptome by high-throughput sequenc-
ing. Nat Genet 40, 1413–1415.
15. Wang, E. T., Sandberg, R., Luo, S., Khreb-
tukova, I., Zhang, L., Mayr, C., Kingsmore,
S.F.,Schroth,G.P.,Burge,C.B.
(2008) Alternative isoform regulation in
human tissue transcriptomes. Nature 456,
470–476.
16. Ji, Z., Lee, J. Y., Pan, Z., Jiang, B., Tian,
B. (2009) Progressive lengthening of 3
untranslated regions of mRNAs by alterna-
tive polyadenylation during mouse embry-
onic development. Proc Natl Acad Sci USA
106, 7028–7033.
17. Sandberg, R., Neilson, J. R., Sarma, A.,
Sharp, P. A., Burge, C. B. (2008) Prolifer-
ating cells express mRNAs with shortened
3
untranslated regions and fewer microRNA
target sites. Science 320, 1643–1647.
18. Moore, M. J. (2005) From birth to death:
the complex lives of eukaryotic mRNAs. Sci-
ence 309, 1514–1518.
19. Diehn, M., Bhattacharya, R., Botstein, D.,
Brown, P. O. (2006) Genome-scale iden-
tification of membrane-associated human
mRNAs. PLoS Genet 2, e11.
20. Halbeisen, R. E., Galgano, A., Scherrer,
T., Gerber, A. P. (2008) Post-transcriptional
gene regulation: from genome-wide studies
to principles. Cell Mol Life Sci 65, 798–813.
21. St Johnston, D. (2005) Moving messages:
the intracellular localization of mRNAs. Nat
Rev Mol Cell Biol 6, 363–375.
22. Gilbert, W. V., Zhou, K., Butler, T. K.,
Doudna, J. A. (2007) Cap-independent
translation is required for starvation-
induced differentiation in yeast. Science 317,
1224–1227.
23. Ingolia, N. T., Ghaemmaghami, S., New-
man, J. R., Weissman, J. S. (2009) Genome-
wide analysis in vivo of translation with
nucleotide resolution using ribosome profil-
ing. Science 324, 218–223.
24. Keene, J. D., Komisarow, J. M., Frieders-
dorf, M. B. (2006) RIP-Chip: the isolation
and identification of mRNAs, microRNAs
and protein components of ribonucleopro-
tein complexes from cell extracts. Nat Protoc
1, 302–307.
25. Baroni, T. E., Chittur, S. V., George, A.
D., Tenenbaum, S. A. (2008) Advances
in RIP-chip analysis: RNA-binding pro-
tein immunoprecipitation-microarray profil-
ing. Methods Mol Biol 419, 93–108.
26. Tian, Q., Stepaniants, S. B., Mao, M., Weng,
L.,Feetham,M.C.,Doyle,M.J.,Yi,E.C.,
Dai, H., Thorsson, V., Eng, J., et al. (2004)
Integrated genomic and proteomic analyses
of gene expression in Mammalian cells. Mol
Cell Proteomics 3, 960–969.
27. Lunde, B. M., Moore, C., Varani, G. (2007)
RNA-binding proteins: modular design for
efficient function. Nat Rev Mol Cell Biol 8,
479–490.
28. Gerber, A. P., Herschlag, D., Brown, P. O.
(2004) Extensive association of functionally
and cytotopically related mRNAs with Puf
family RNA-binding proteins in yeast. PLoS
Biol 2, E79.
29. Saint-Georges, Y., Garcia, M., Delaveau, T.,
Jourdren, L., Le, C. S., Lemoine, S., Tanty,
V., Devaux, F., Jacq, C. (2008) Yeast mito-
chondrial biogenesis: a role for the PUF
RNA-binding protein Puf3p in mRNA local-
ization. PLoS One 3, e2293.
30. Townley-Tilson, W. H., Pendergrass, S. A.,
Marzluff, W. F., Whitfield, M. L. (2006)
Genome-wide analysis of mRNAs bound to
the histone stem-loop binding protein. RNA
12, 1853–1867.
31. Barreau, C., Paillard, L., Osborne, H. B.
(2005) AU-rich elements and associated fac-
tors: are there unifying principles? Nucleic
Acids Res 33, 7138–7150.

Chapter 17
RIP-Chip Analysis:
R
NA-Binding Protein
I
mmuno
p
recipitation-Microarray (
Chip
) Profiling
Ritu Jain, Tiffany Devine, Ajish D. George, Sridar V. Chittur,
Timothy E. Baroni, Luiz O. Penalva, and Scott A. Tenenbaum
Abstract
Post-transcriptional regulation of gene expression plays an important role in complex cellular processes.
Just like transcription factors regulate gene expression through combinatorial binding to multiple, phys-
ically dispersed cis elements, mRNA binding proteins can regulate the translation of functionally related
gene products by coordinately binding to subsets of mRNAs. The networks of mRNA binding proteins
that facilitate this fine-tuning of gene expression are poorly understood. By combining genomic technolo-
gies with standard molecular biology tools, we have helped pioneer the development of high-throughput
technologies for the global analysis of subsets of mRNAs bound to RNA-binding proteins. This tech-
nique is termed RIP-Chip and stands for RNA-Binding Protein Immunoprecipitation-Microarray (Chip)
Profiling. This approach is also referred to as “ribonomic profiling” and has revealed valuable information
about the workings of mRNP networks in the cell and the regulation of gene expression. In this chapter,
we describe the latest advances that we have made in the RIP-CHIP technology.
Key words: Post-transcriptional gene regulation, Ribonomics, RIP-Chip, RNA-binding Pro-
tein (RBP), Immunoprecipitation (IP), microarray, microarray expression profiling, array, systems
biology.
1. Introduction
In eukaryotic organisms, a tight regulation of gene expression
is required for complex processes such as development, differen-
tiation, cell specification, and response of each cell to environ-
mental signals. Any deviation from this regulation may result in
disease and other abnormal conditions. The multistep process
of translating the information contained in the genetic code of
H. Nielsen (ed.), RNA, Methods in Molecular Biology 703,
DOI 10.1007/978-1-59745-248-9_17, © Springer Science+Business Media, LLC 2011
247
248 Jain et al.
every cell into proteins that ultimately control the function of
the cell is regulated at every step (for a review, see 1). Thus far,
scientists have extensively studied the beginning (transcription),
middle (splicing), and the end (translation and post-translational
modifications) of this process. However, it has been well docu-
mented that there is a discrepancy between mRNA transcript lev-
els and the final amount of protein produced. There is increasing
evidence that RNA does not exist as a naked entity in the cell
but is always bound by proteins that guide it through every step.
All events beginning with the transcription of mRNA to process-
ing, transport, localization, stability, degradation, and translation
are regulated by various RNA-Binding Proteins (RBPs), and con-
tribute significantly to the final amount of protein product pro-
duced (for a review, see 2–4). These RNA binding proteins are
required to accomplish several complicated tasks such as localiza-
tion of mRNAs during embryonic patterning (5), development
of synaptic plasticity (6, 7), maintaining iron homeostasis in the
cell (8), response to growth signals (9) and regulation of cellu-
lar growth (10). The complex of mRNA and its associated set of
proteins and small non-coding RNAs is commonly referred to as
an mRNP (messenger ribonucleoprotein particle). The set of pro-
teins associated with mRNA may change dynamically and all these
processes are coordinated to generate a gene expression profile
specific for that cell under a particular condition.
In eukaryotes, the process of transcription is physically sep-
arated from translation by the nuclear membrane. In addition
the functionally related genes are dispersed on the chromo-
some. However, coordinated expression of functionally related
genes can be achieved through RBPs, which can sequester func-
tionally related mRNAs together. These “post-transcriptional
operons” may allow the cell to generate a rapid response to an
incoming signal by regulating the stability and translation of spe-
cific mRNAs (11). The post-transcriptional oper on model can be
used to explain the discordance that is often observed between the
levels of mRNA and the protein of any given gene. Just because
a gene is transcribed does not necessarily mean that it is trans-
lated. It may be kept on hold, or degraded by various RBPs.
Experimental evidence from many different labs in recent years
has demonstrated that mRNAs which either participate in a com-
mon function or subcellular localization tend to be part of the
same RNP complex, thus providing further evidence for the exis-
tence of post-transcriptional operons. Often, these RBPs bind to
mRNAs through elements that contain sequence and structural
specificity and are present in their 5
or 3
untranslated regions.
Each mRNA often contains binding-sites for more than one RBP
and each RBP has the potential to associate with more than one
mRNA, giving rise to combinatorial, systems-level regulatory net-
works with the potential for tremendous complexity, yet elegant
RIP-Chip Analysis 249
simplicity (12). This combinatorial post-transcriptional network-
ing likely gives rise to much of the complexity of the human
genome that has yet to be fully understood. RIP-Chip provides
the fundamental technology to elucidate this information.
The complete human genome sequence has revealed the
predicted pr e sence of 1,000–2,000 RBPs. Each of these RBPs
may bind subsets of 10–1,000 s of mRNAs at a time. Individ-
ual analysis of each of these mRNAs bound to their respective
proteins one-at-a-time would be painstakingly slow and labori-
ous. To facilitate this, we have helped pioneer the development
of high-throughput technologies to analyze the entire subset of
mRNAs associated with a particular RBP at a global level, using
microarrays (12–15). This technique which employs immunopre-
cipitation of endogenously formed mRNP complexes using an
antibody specific to the RBP in question, followed by purifica-
tion of associated mRNAs and their global, quantitative analysis
using a genomic read-out such as microarrays, SAGE analysis, or
deep-sequencing has been ter med RIP-Chip (RNA-Binding Pro-
tein Immunoprecipitation-Microarray (Chip)), and is outlined in
Fig. 17.1. Many refer to the approach of characterizing mRNA
subsets associated with mRNA binding proteins as “ribonomics,”
and this approach has been widely used to identify unique mRNA
regulatory networks targeted by various mRNA binding proteins
(13–57). Since this method can also detect mRNAs that do not
directly bind to a particular RBP but may form part of the larger
mRNP complex, this method also provides a way to determine
the mRNA infrastructure of a cell or tissue. Ribonomic profil-
ing has provided several valuable insights into post-transcriptional
gene networks, e.g., we were able to show that upon functional
perturbation (stimulation of P19 embryonic carcinoma cells with
Retinoic Acid, stress, differentiation, etc.), the subset of mRNAs
associated with any particular RBP may change (13). Compari-
son of mRNAs associated with the Fragile-X Mental Retardation
protein (FMRP) from normal and Fragile-X syndrome patients
revealed the subset of mRNAs whose translation may change
in the absence of FMRP (16). Also, upon phosphorylation, the
localization of the La protein and its associated RNAs were
altered (18).
In the last few years many groups have used RIP-
Chip profiling to study the mRNA profiles associated with
selected RBPs (12–58). Some have used different meth-
ods to co-immunoprecipitate mRNAs bound to a particu-
lar RBP to further analyze the mRNA targets (e.g., cloning
and characterization, high-throughput sequencing, etc.). As a
further modification of the technique, groups have ectopi-
cally expressed the RBP as a recombinant fusion pro-
tein (e.g., Flag-tag or TAP-tag), and performed RIP-Chip
using antibodies against the tag. Several have employed UV
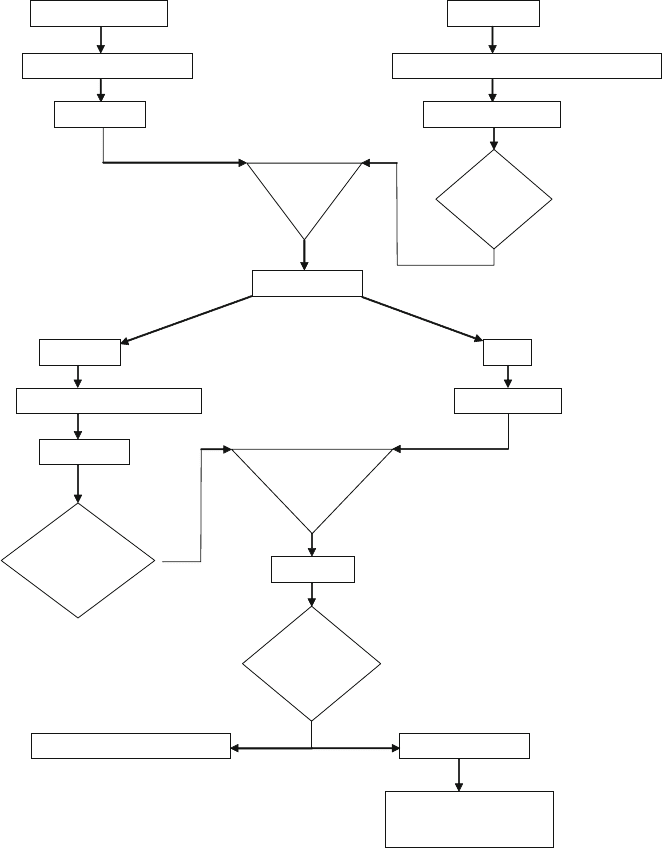
250 Jain et al.
Swell beads (A or G)
Bind antibody to the beads
Wash beads
Harvest cells
Prepare Lysate with Polysome Lysis Buffer
Spin down the lysate
Add lysateto
the beads
Take out aliquot
IP reaction
Input
Incubate at 4
0
C with tumbling Incubate at 4
0
C
Wash beads
Check
efficiency of
immuno-
precipitation by
Western
Proteinase K
treatment at 55
0
C
for 30 min.
Purify RNA
Are the IP
and Input
RNA of good
quality?
RT-PCR to look for targets in IP
Microarray analysis
Look for enrichment of
targets in IP vs. Input and
(-) control
Is the lysate
of good
quality?
Fig. 17.1. Flow-chart for the RIP-CHIP technique.
cross-linking to irreversibly bind mRNA targets to RBP, a tech-
nique known as CLIP (40, 41). Collectively of these studies
have provided valuable insights into the function of mRNPs. For
example, a systematic analysis of mRNAs bound to 40 different
RBPs of yeast has revealed that each binds a subset of mRNAs
with common functional or subcellular localization themes (28).
These subsets may overlap indicating a combinatorial network of
regulation. Ribonomic profiling has been extended to include
analysis of miRNA regulation of translation. Immunopurifica-
tion of miRNP has revealed valuable information about miRNA

RIP-Chip Analysis 251
and their mRNA targets (33–38). Interestingly, many well-known
RBPs (e.g., HuR, FMRP, Staufen) were found to be associated
with miRNP complexes providing evidence for extensive connec-
tions between RBP and miRNA regulatory networks ( 27, 38).
Immunopurification-based methods for identifying miRNA tar-
gets has the potential to be more biologically relevant than com-
puter prediction-based approaches because it allows the direct
identification of miRNA targets with non-ideal sequence com-
plementarity and without any prior knowledge of the rules of
interaction between miRNA and its targets. Lastly, ribonomic
profiling has also been used to identify mRNA targets of RBPs
from microorganisms such as Salmonella and Escherichia coli
(43, 44).
In this chapter, we describe the most current RIP-Chip tech-
niques as we presently practice them and provide helpful tips and
suggestions to present a comprehensive overview of the technol-
ogy in hopes that it can be as widely used as possible.
2. Materials
We recommend that throughout this method, all standard pre-
cautions should be taken to minimize RNAse contamination (see
Notes 1 and 2). All reagents, glassware, and plasticware should
either be purchased RNAse-DNAse free or be treated with 0.1%
DEPC to ensure that they are RNAse free. All cell culture should
be done with cell-culture grade media.
1. Tissues and/or cells of interest.
2. Phosphate-buffered Saline (PBS).
3. Bovine Serum Albumin Fraction V.
4. Antibody to the RNA Binding Protein of interest.
5. Antibody binding matrix (e.g., Protein A Sephar ose
(Sigma) or Protein G Sepharose beads (Sigma), or the
pre-swollen beads from GE Healthcare, or Magnetic beads
(Dynabeads) from Invitrogen). Choice of beads depends
on the antibody being used (see Notes 3 and 4).
6. 0.1 M sodium phosphate buffer (pH 8.1) for Dynabeads
A or 0.1 M citrate-phosphate buffer (pH 5.0) for Dyn-
abeads G.
7. 0.2 M triethanolamine (Sigma).
8. 20 mM DMP (dimethyl pimelimidate (Sigma)) in 0.2 M
triethanolamine (freshly prepared).
9. 0.1 M DTT.

252 Jain et al.
10. 0.5 M EDTA, pH 8.0.
11. 5 M ammonium acetate.
12. 7.5 M lithium chloride.
13. Glycogen, 5 mg/mL (Ambion).
14. Proteinase K, 20 mg/mL (Ambion).
15. Acid-phenol:chloroform, pH 4.5 (with isoamyl alcohol,
25:24:1) (Ambion).
16. Chloroform.
17. Absolute ethanol.
18. 80% ethanol.
19. RNAse OUT, 40 U/mL (Invitrogen) or RNAse Inhibitor
(New England Biolabs) or Superase•IN
TM
(Ambion, 20
U/μL).
20. Complete
TM
proteinase inhibitor cocktail (Roche).
21. Nuclease-free water (Ambion).
We recommend using nuclease-free water to prepare the fol-
lowing buffers:
1. Polysome Lysis Buffer (PLB): 10 mM Hepes, pH 7.0,
100 mM KCl, 5 mM MgCl
2
, 0.5% NP-40.
Typically we prepare a 10× stock of this buffer from
which a ready to use Polysome Lysis Buffer can be prepared.
At the time of use, 1 mM DTT, 100 Units/mL R NAse
OUT, and complete protease inhibitor cocktail should be
added (see Note 5).
2. NT-2 Buffer: 50 mM Tris-HCl, pH 7.4, 150 mM NaCl,
1mMMgCl
2
, 0.05% NP-40. Typically we prepare a 5×
stock of NT-2 Buffer, from which a ready to use buffer can
be prepared as and when desired.
3. NET-2 Buffer (Binding buffer for RNP to antibody): NT-2
buffer supplemented with 20 mM EDTA, pH 8.0, 1 mM
DTT, 100 U/mL RNAse OUT.
4. Proteinase K digestion buffer: NT-2 buffer supplemented
with 1% SDS, 1.2 mg/mL Proteinase K.
5. Antibody binding buffer: 5% BSA in NT-2 buffer.
3. Methods
The method described here has been successfully used with mam-
malian cells and tissue samples, as well as Caenorhabditis elegans
and Drosophila melanogaster cells. However, we assume that this
method could be adapted or modified to samples fr om other
RIP-Chip Analysis 253
organisms. A flow-chart explaining the various steps involved in
RIP-Chip is shown in Fig. 17.1. We have successfully immuno-
precipitated RNA Binding Protein-mRNA complexes (RIP) using
both Sepharose and magnetic beads. Both methods are detailed
here. Typically immunoprecipitations are performed in a 1 mL
volume, this allows for reproducible kinetics, reduced mRNA re-
assortment potential, and ease of calculations.
3.1. Preparation of
PLB Lysates
1. Grow desired cells to about 70–80% confluence, on 150- or
100-mm plates.
2. Wash twice with ice-cold PBS.
3. Harvest cells by scraping in ice-cold PBS and transfer to a
centrifuge tube. If desired, Trypan Blue assay can be used to
check the viability of the cells at this point.
4. Collect cells by centrifugation at 2,500×g for 5 min.
5. Aspirate PBS from the cell pellet and add equal pellet vol-
ume of PLB (containing protease and RNAse inhibitors) to
the cell pellet. Mix by pipetting up and down until the mix-
ture looks homogeneous. Keep on ice for 5 min to allow
the hypotonic PLB buffer to swell the cells. Dispense into
1-mL aliquots and store at –80
◦
C. This method depends on
repeated freeze–thaws to gently lyse the cells. The cell lysates
are ready to proceed to RBP immunoprecipitation (see
Note 6).
3.2. Coating of Bead
Matrix with Antibody
3.2.1. Coating
Sepharose Beads
1. Swell the desired amount of protein G or protein A agarose
beads in NT2 buffer containing 5% BSA. Typically we add
30 mL of buffer to 1 g of beads. This yields 20% slurry (1 mL
of slurry yields about 200 μL of packed bead volume). Alter-
natively, pre-swollen beads equilibrated with NT2 buffer can
be used.
2. Add the immunoprecipitation antibody or serum and incu-
bate for at least 1 h at room temperature on a rotating device
or overnight (or longer) at 4
◦
C(see Notes 7 and 8). Typi-
cally for each immunoprecipitation reaction we use 300 μL
of slurry, dilute it with 1 mL of NT2 buffer containing 5%
BSA, and add 5 μg of commercially pur chased antibody.
3.2.2. Coating Magnetic
Beads
For both Dynabeads protein A and protein G, the method is
essentially as recommended by the manufacturer.
1. For each immunoprecipitation reaction use 50 μLof
Dynabeads A suspension, wash twice with 0.5 mL of
0.1 M sodium phosphate buffer (pH 8.1) using the mag-
net Dynal. For Dynabeads G use citrate-phosphate buffer
