Arora R. (ed.) Medicinal Plant Biotechnology
Подождите немного. Документ загружается.

234 Analytical Platforms and Databases
users can obtain structural information for peaks detected during metabolic profiling
without the need for performing additional MS/MS analysis; structural information can be
obtained by searching for the corresponding MS2T accession in the libraries.
Pathway databases
In order to understand the molecular basis of life, it is important to clarify the relationships
between cells, organisms and the biosphere. Hence, in the recent years, interactions
between and pathways involving genes and molecules are rapidly being organized after
which protein activities, gene annotations and metabolites will be integrated into a single
database. Some databases for metabolic pathways are listed in Table 14.8.
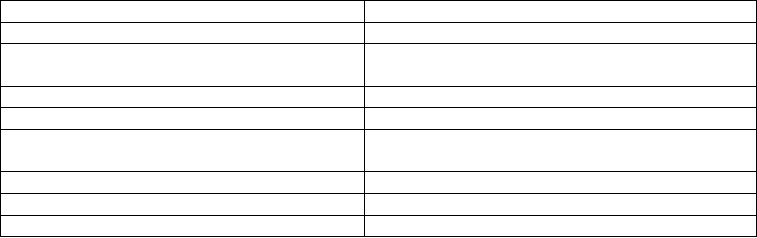
Table 14.8. Pathway databases.
AraCyc at TAIR http://www.Arabidopsis.org/biocyc/index.jsp
PlantCyc http://plantcyc.org/
Kyoto Encyclopedia of Genes and
Genomes (KEGG) Pathway Database
http://www.genome.jp/kegg/pathway.html
MetaCrop http://metacrop.ipk-gatersleben.de/
MapMan http://gabi.rzpd.de/projects/MapMan/
Kazusa Plant Pathway Viewer (KaPPA-
View)
http://kpv.kazusa.or.jp/kpv3/guestIndex.jsp
Metabolic Pathway Search at KATANA http://www.kazusa.or.jp/katana/pathway.html
MetaCyc http://metacyc.org/
BioCyc http://biocyc.org/
AraCyc
http://www.Arabidopsis.org/biocyc/
AraCyc (Mueller et al., 2003) provides access to manually curated or reviewed information
on metabolic pathways in Arabidopsis– the model plant. These pathways may be unique to
Arabidopsis or common to other organisms. Presently, this database features more than 170
pathways, and includes information on compounds, intermediates, cofactors, reactions,
genes, proteins, and subcellular locations of proteins. The database uses the Pathway Tools
software (Karp et al., 2002), which allows users to visualize all pathways in the database
down to the individual chemical structures of the compounds.
PlantCyc of the Plant Metabolic Network
http://plantcyc.org/
PlantCyc contains pathways and the catalytic enzymes and genes involved in pathways as
well as compounds from many plant species. Majority of the pathway diagrams in PlantCyc
were manually extracted from the plant literature. They are either supported by
experimental evidence or are based on expert hypotheses. A small portion of the pathways
were computationally predicted. These pathways are known to exist in non-plant organisms
but have not been found in plants. However, some hints indicate that plants may utilize the
same pathway or similar routes. Both experimentally verified and computationally
predicted enzymes and genes are assigned to pathways. Evidence codes are designated to
the pathways and genes/enzymes are assigned in order to provide information on data
Analytical Platforms and Databases 235
quality. In addition to new pathways curated in-house, the first release of PlantCyc contains
all pathways from AraCyc (Mueller et al., 2003), all plant pathways from MetaCyc (Caspi
et al., 2008), and a number of curated pathways from other plant pathway databases
including RiceCyc (Jaiswal et al., 2006) and MedicCyc (Urbanczyk-Wochniak and Sumner,
2007). Moreover PlantCyc integrates computational assignments of enzymes and genes
from other plant pathway databases including AraCyc, RiceCyc, MedicCyc, and SolaCyc.
KEGG PATHWAY
http://www.genome.jp/kegg/pathway.html
The KEGG pathway contains information on metabolites and genes as well as graphical
representations of metabolic pathways and complexes derived from various biological
processes (Kanehisa et al., 2002). Thus far, 94,739 pathways generated from 327 reference
pathways for over 200 organisms, including Arabidopsis and rice, have been constructed.
Maps of organism-specific metabolic pathways can be generated on the basis of the
assignment information in the KEGG/GENES database.
MetaCrop
http://metacrop.ipk-gatersleben.de/
MetaCrop (Grafahrend-Belau et al., 2008) is a database containing manually curated,
highly detailed information on metabolic pathways in crop plants; this database includes
location information as well as information on transport processes and reaction kinetics.
The web interface enables an easy exploration of information from overview pathways to
single reactions, and thus helps users understand the metabolism of crop plants. It also
supports automatic data export for the creation of detailed models of metabolic pathways in
order to support simulation approaches. MetaCrop contains hand-curated information on
approximately 40 major metabolic pathways in various crop plants; in particular, the
metabolism of agronomically important organs such as seed or tuber is available. In this
database, species of both monocotyledons and dicotyledons are represented. Information on
involved metabolites, stoichiometry, and the precise location (species, organ, tissue,
compartment, and developmental stage) has been incorporated in the reactions.
Furthermore, with regard to central metabolism (glycolysis, TCA cycle), kinetic data of the
reactions are available. References and relevant PubMed IDs are provided. In order to have
a controlled vocabulary for allowing the comparison of data from different sources,
ontology terms have been used. In total, the database currently contains information for six
major crop plants (approximately 400 enzymatic reactions, 60 transport processes, 6
compartments, and 600 references) and initial information for several more crop plants. All
information was manually extracted through an extensive survey of literature and online
databases.
MapMan
http://gabi.rzpd.de/projects/MapMan/
MapMan (Thimm et al., 2004; Usadel et al., 2005) is a user-driven tool that displays large
genomics datasets onto diagrams of metabolic pathways or other processes. MapMan
consists of a Scavenger module that collects and classifies the measured parameters into
hierarchical functional categories, and an ImageAnnotator module that uses these
236 Analytical Platforms and Databases
classifications to organize and display data on diagrams of the user's choice. Each gene is
represented by a discrete signal, allowing individual responses to be identified. Signals for
genes that are involved in a particular process are grouped spatially, making it possible to
discern general trends that would be less apparent from lists of individual genes. Now,
users can browse more than a dozen sample data sets. Moreover the MapMan has been
distributed as a JAVA application, therefore, users can perform their analyses by using
their own PCs.
Kazusa Plant Pathway Viewer
http://kpv.kazusa.or.jp/kappa-view/
With advances in DNA array technology, vast amounts of transcriptome data have been
produced, and mass spectrometers have contributed to the massive production of
metabolome data. Interpretation of metabolome data in conjunction with transcriptome data
is one of the major concerns associated with research on the identification of metabolism-
related gene functions. Kazusa Plant Pathway Viewer (KaPPA-View) (Tokimatsu et al.,
2005) facilitates the representation of quantitative data for individual transcripts and/or
metabolites on plant metabolic pathway maps, and easy comprehension of the transcripts
and metabolites; this in turn facilitates investigation into the hypotheses of gene functions
in metabolic pathways.
Assembly of Transcriptomics and Metabolomics
In order to understand the mechanisms underlying the functioning of a living organism by
linking a range of information from the genome, transcriptome, proteome and metabolome
of the organism, it is essential to have a platform that is capable of integrating a range of
‘omics’ data so that these data can be analysed together. ‘Batch-learning self-organizing
map’ (BL-SOM) is one of the tools that can support such integrated analyses (Kanaya et al.,
2001; Abe et al., 2003). The transcriptome and metabolome datasets of this tool consist of
a matrix in which signal intensities are arranged in multiple columns (experimental series)
and multiple rows (gene and metabolite IDs). BL-SOM can analyse an integrated matrix of
both transcriptome and metabolome data after appropriate normalization of the data and
preliminary calculations; thus, this tool allows researchers to visualize the correlations
between elements. Genes and metabolites are classified into clusters in a two-dimensional
‘feature map’ based on the expression and accumulation patterns of genes and metabolites.
The high reproducibility and resolution supported by BL-SOM enable the elucidation of
gene-to-metabolite, gene-to-gene and metabolite-to-metabolite correlations, which facilitate
the identification of a gene’s function (Hirai et al., 2004, 2005).
One example stated above is PRIMe (Akiyama et al., 2008). PRIMe is a web site that
has been designed and implemented to support research and analysis workflows ranging
from metabolome to transcriptome analysis. This site provides access to a growing
collection of standardized measurements of metabolites obtained by using NMR, GC-MS,
LC-MS, CE-MS, and metabolomics tools that support related analyses (SpinAssign for the
identification of metabolites by means of NMR, and KNApSAcK (Shinbo et al., 2006) for
searches within metabolite databases). In addition, the transcriptomics tools support
Correlated Gene Search, and Cluster Cutting for the analysis of mRNA expression. The use
of these tools and databases can contribute to the analysis of biological events at the level

Analytical Platforms and Databases 237
of metabolite and gene expression. For Arabidopsis thaliana, we describe one example of
such an analysis using BL-SOM (Kanaya et al., 2001), which can be accessed from the
web site.
Conclusion
Because of space constraints, we were unable to introduce all the existing databases and
tools; however, several hundreds of information resources for plant research have been
generated and are being updated. This implies that our research environment is also
improving. We can reduce the cost of research activities and efficiently focus on further
advancements in our studies, which will generate newly analysed results. In other words,
new information resources will be more enriched, and will certainly accelerate research in
the field of plant sciences. The recent technological advancement in the proteome and
metabolome research areas is remarkable. We strongly feel that there have been increasing
advances in studies on proteomics and metabolomics. Similarly, there is an increasing need
for the study of phenomics. Further, we should consider using new integrative techniques
and methods for our future research. If advancements in these are successful, we may
obtain further insights into cells and life systems.
References
Abe, T., Kanaya, S., Kinouchi, M., Ichiba, Y., Kozuki, T. and Ikemura, T. (2003) Informatics for
unveiling hidden genome signatures. Genome Research 13, 693–702.
Akiyama, K., Chikayama, E., Yuasa, H., Shimada, Y., Tohge, T., Shinozaki, K., Hirai, M.Y., Sakurai,
T., Kikuchi, J. and Saito, K. (2008) PRIMe: a web site that assembles tools for metabolomics and
transcriptomics. In silico Biology 8, 339–345.
Aoki, K., Ogata, Y. and Shibata, D. (2007) Approaches for extracting practical information from
gene co-expression networks in plant biology. Plant & Cell Physiology 48, 381–390.
Axtell, M.J., Jan, C., Rajagopalan, R. and Bartel, D.P. (2006) A two-hit trigger for siRNA biogenesis
in plants. Cell 127, 565–577.
Barrett, T., Troup, D.B., Wilhite, S.E., Ledoux, P., Rudnev, D., Evangelista, C., Kim, I.F., Soboleva,
A., Tomashevsky, M., Marshall, K.A., Phillippy, K.H., Sherman, P.M., Muertter, R.N. and Edgar,
R. (2009) NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids
Research 37, D885–890.
Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M.C., Estreicher, A., Gasteiger, E., Martin, M.J.,
Michoud, K., O'Donovan, C., Phan, I., Pilbout, S. and Schneider, M. (2003) The SWISS–PROT
protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Research 31, 365–
370.
Bottcher, C., von Roepenack-Lahaye, E., Schmidt, J., Schmotz, C., Neumann, S., Scheel, D. and
Clemens, S. (2008) Metabolome analysis of biosynthetic mutants reveals a diversity of metabolic
changes and allows identification of a large number of new compounds in Arabidopsis. Plant
Physiology 147, 2107–2120.
Brodersen, P., Sakvarelidze-Achard, L., Bruun-Rasmussen, M., Dunoyer, P., Yamamoto, Y.Y.,
Sieburth, L. and Voinnet, O. (2008) Widespread translational inhibition by plant miRNAs and
siRNAs. Science 320, 1185–1190.
Caspi, R., Foerster, H., Fulcher, C.A., Kaipa, P., Krummenacker, M., Latendresse, M., Paley, S.,
Rhee, S.Y., Shearer, A.G., Tissier, C., Walk, T.C., Zhang, P. and Karp, P.D. (2008) The MetaCyc
238 Analytical Platforms and Databases
Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome
Databases. Nucleic Acids Research 36, D623–631.
Claros, M.G. and Vincens, P. (1996) Computational method to predict mitochondrially imported
proteins and their targeting sequences. European Journal of Biochemistry/FEBS 241, 779–786.
Craigon, D.J., James, N., Okyere, J., Higgins, J., Jotham, J. and May, S. (2004) NASCArrays: a
repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids
Research 32, D575–577.
Cui, J., Li, P., Li, G., Xu, F., Zhao, C., Li, Y., Yang, Z., Wang, G., Yu, Q., Li, Y. and Shi, T. (2008a)
AtPID: Arabidopsis thaliana protein interactome database-an integrative platform for plant
systems biology. Nucleic Acids Research 36, D999–1008.
Cui, Q., Lewis, I.A., Hegeman, A.D., Anderson, M.E., Li, J., Schulte, C.F., Westler, W.M.,
Eghbalnia, H.R., Sussman, M.R. and Markley, J.L. (2008b) Metabolite identification via the
Madison Metabolomics Consortium Database. Nature Biotechnology 26, 162–164.
Dettmer, K., Aronov, P.A. and Hammock, B.D. (2007) Mass spectrometry-based metabolomics.
Mass Spectrometry Reviews 26, 51–78.
Dixon, R.A., Gang, D.R., Charlton, A.J., Fiehn, O., Kuiper, H.A., Reynolds, T.L., Tjeerdema, R.S.,
Jeffery, E.H., German, J.B., Ridley, W.P. and Seiber, J.N. (2006) Applications of metabolomics
in agriculture. Journal of Agricultural and Food Chemistry 54, 8984–8994.
Eisen, M.B., Spellman, P.T., Brown, P.O. and Botstein, D. (1998) Cluster analysis and display of
genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United
States of America 95, 14863–14868.
Emanuelsson, O., Brunak, S., von Heijne, G. and Nielsen, H. (2007) Locating proteins in the cell
using TargetP, SignalP and related tools. Nature Protocols 2, 953–971.
Fargione, J., Hill, J., Tilman, D., Polasky, S. and Hawthorne, P. (2008) Land clearing and the biofuel
carbon debt. Science 319, 1235–1238.
Fiehn, O. (2002) Metabolomics–the link between genotypes and phenotypes. Plant Molecular
Biology 48, 155–171.
Fiehn, O., Kloska, S. and Altmann, T. (2001) Integrated studies on plant biology using multiparallel
techniques. Current Opinion in Biotechnology 12, 82–86.
Friso, G., Giacomelli, L., Ytterberg, A.J., Peltier, J.B., Rudella, A., Sun, Q. and Wijk, K.J. (2004) In-
depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new
proteins, new functions, and a plastid proteome database. The Plant Cell 16, 478–499.
Galperin, M.Y. (2008) The Molecular Biology Database Collection: 2008 update. Nucleic Acids
Research 36, D2–4.
Galperin, M.Y. and Cochrane, G.R. (2009) Nucleic Acids Research annual Database Issue and the
NAR online Molecular Biology Database Collection in 2009. Nucleic Acids Research 37, D1–4.
Gandhi, T.K., Zhong, J., Mathivanan, S., Karthick, L., Chandrika, K.N., Mohan, S.S., Sharma, S.,
Pinkert, S., Nagaraju, S., Periaswamy, B., Mishra, G., Nandakumar, K., Shen, B., Deshpande, N.,
Nayak, R., Sarker, M., Boeke, J.D., Parmigiani, G., Schultz, J., Bader, J.S. and Pandey, A. (2006)
Analysis of the human protein interactome and comparison with yeast, worm and fly interaction
datasets. Nature Genetics 38, 285–293.
Geisler-Lee, J., O'Toole, N., Ammar, R., Provart, N.J., Millar, A.H. and Geisler, M. (2007) A
predicted interactome for Arabidopsis. Plant Physiology 145, 317–329.
Giot, L., Bader, J.S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y.L., Ooi, C.E., Godwin,
B., Vitols, E., Vijayadamodar, G., Pochart, P., Machineni, H., Welsh, M., Kong, Y., Zerhusen, B.,
Malcolm, R., Varrone, Z., Collis, A., Minto, M., Burgess, S., McDaniel, L., Stimpson, E., Spriggs,
F., Williams, J., Neurath, K., Ioime, N., Agee, M., Voss, E., Furtak, K., Renzulli, R., Aanensen,
N., Carrolla, S., Bickelhaupt, E., Lazovatsky, Y., DaSilva, A., Zhong, J., Stanyon, C.A., Finley,
R.L., Jr., White, K.P., Braverman, M., Jarvie, T., Gold, S., Leach, M., Knight, J., Shimkets, R.A.,
McKenna, M.P., Chant, J. and Rothberg, J.M. (2003) A protein interaction map of Drosophila
melanogaster. Science 302, 1727–1736.
Goda, H., Sasaki, E., Akiyama, K., Maruyama–Nakashita, A., Nakabayashi, K., Li, W., Ogawa, M.,
Yamauchi, Y., Preston, J., Aoki, K., Kiba, T., Takatsuto, S., Fujioka, S., Asami, T., Nakano, T.,
Analytical Platforms and Databases 239
Kato, H., Mizuno, T., Sakakibara, H., Yamaguchi, S., Nambara, E., Kamiya, Y., Takahashi, H.,
Hirai, M.Y., Sakurai, T., Shinozaki, K., Saito, K., Yoshida, S. and Shimada, Y. (2008) The
AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation,
model data analysis and data access. Plant Journal 55, 526–542.
Grafahrend-Belau, E., Weise, S., Koschutzki, D., Scholz, U., Junker, B.H. and Schreiber, F. (2008)
MetaCrop: a detailed database of crop plant metabolism. Nucleic Acids Research 36, D954–958.
Gustafson, A.M., Allen, E., Givan, S., Smith, D., Carrington, J.C. and Kasschau, K.D. (2005) ASRP:
the Arabidopsis Small RNA Project Database. Nucleic Acids Research 33, D637–640.
Heazlewood, J.L., Tonti-Filippini, J.S., Gout, A.M., Day, D.A., Whelan, J. and Millar, A.H. (2004)
Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and
regulatory components, provides assessment of targeting prediction programs, and indicates
plant-specific mitochondrial proteins. The Plant Cell 16, 241–256.
Heazlewood, J.L., Tonti-Filippini, J., Verboom, R.E. and Millar, A.H. (2005) Combining
experimental and predicted datasets for determination of the subcellular location of proteins in
Arabidopsis. Plant Physiology 139, 598–609.
Heazlewood, J.L., Verboom, R.E., Tonti-Filippini, J., Small, I. and Millar, A.H. (2007) SUBA: the
Arabidopsis Subcellular Database. Nucleic Acids Research 35, D213–218.
Hirai, M.Y., Yano, M., Goodenowe, D.B., Kanaya, S., Kimura, T., Awazuhara, M., Arita, M.,
Fujiwara, T. and Saito, K. (2004) Integration of transcriptomics and metabolomics for
understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proceedings of
the National Academy of Sciences of the United States of America 101, 10205–10210.
Hirai, M.Y., Klein, M., Fujikawa, Y., Yano, M., Goodenowe, D.B., Yamazaki, Y., Kanaya, S.,
Nakamura, Y., Kitayama, M., Suzuki, H., Sakurai, N., Shibata, D., Tokuhisa, J., Reichelt, M.,
Gershenzon, J., Papenbrock, J. and Saito, K. (2005) Elucidation of gene-to-gene and metabolite-
to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. The Journal
of Biological Chemistry 280, 25590–25595.
Hooper, S.D. and Bork, P. (2005) Medusa: A simple tool for interaction graph analysis.
Bioinformatics (Oxford, England) 21, 4432–4433.
Horai, H., Arita, M. and Nishioka, T. (2008) Comparison of ESI–MS Spectra in MassBank Database.
Proceedings of the International Conference on BioMedical Engineering and Informatics 853–
857.
Howell, M.D., Fahlgren, N., Chapman, E.J., Cumbie, J.S., Sullivan, C.M., Givan, S.A., Kasschau,
K.D. and Carrington, J.C. (2007) Genome-wide analysis of the RNA–DEPENDENT RNA
POLYMERASE6/DICER–LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and
tasiRNA-directed targeting. The Plant Cell
, 19, 926–942.
Hruz, T., Laule, O., Szabo, G., Wessendorp, F., Bleuler, S., Oertle, L., Widmayer, P., Gruissem, W.
and Zimmermann, P. (2008) A Reference Expression Database for the Meta-Analysis of
Transcriptomes. Advances in Bioinformatics 2008, 420747.
Ito, T., Motohashi, R., Kuromori, T., Mizukado, S., Sakurai, T., Kanahara, H., Seki, M. and
Shinozaki, K. (2002a) A new resource of locally transposed Dissociation elements for screening
gene–knockout lines in silico on the Arabidopsis genome. Plant Physiology 129, 1695–1699.
Ito, T., Motohashi, R. and Shinozaki, K. (2002b) Preparation of transposon insertion lines and
determination of insertion sites in Arabidopsis genome. Methods in Molecular Biology 182, 209–
219.
Ito, T., Motohashi, R., Kuromori, T., Noutoshi, Y., Seki, M., Kamiya, A., Mizukado, S., Sakurai, T.
and Shinozaki, K. (2005) A resource of 5,814 dissociation transposon-tagged and sequence-
indexed lines of Arabidopsis transposed from start loci on chromosome 5. Plant & Cell
Physiology 46, 1149–1153.
Jaiswal, P., Ni, J., Yap, I., Ware, D., Spooner, W., Youens-Clark, K., Ren, L., Liang, C., Zhao, W.,
Ratnapu, K., Faga, B., Canaran, P., Fogleman, M., Hebbard, C., Avraham, S., Schmidt, S.,
Casstevens, T.M., Buckler, E.S., Stein, L. and McCouch, S. (2006) Gramene: a bird's eye view of
cereal genomes. Nucleic Acids Research 34, D717–723.
240 Analytical Platforms and Databases
Johnson, C., Bowman, L., Adai, A.T., Vance, V. and Sundaresan, V. (2007) CSRDB: a small RNA
integrated database and browser resource for cereals. Nucleic Acids Research 35, D829–833.
Jones-Rhoades, M.W., Bartel, D.P. and Bartel, B. (2006) MicroRNAS and their regulatory roles in
plants. Annual Review of Plant Biology 57, 19–53.
Kanaya, S., Kinouchi, M., Abe, T., Kudo, Y., Yamada, Y., Nishi, T., Mori, H. and Ikemura, T.
(2001) Analysis of codon usage diversity of bacterial genes with a self-organizing map (SOM):
characterization of horizontally transferred genes with emphasis on the E. coli O157 genome.
Gene 276, 89–99.
Kanehisa, M., Goto, S., Kawashima, S. and Nakaya, A. (2002) The KEGG databases at GenomeNet.
Nucleic Acids Research 30, 42–46.
Karp, P.D., Paley, S. and Romero, P. (2002) The Pathway Tools software. Bioinformatics (Oxford,
England) 18 Suppl 1, S225–232.
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., D'Angelo, C., Bornberg-
Bauer, E., Kudla, J. and Harter, K. (2007) The AtGenExpress global stress expression data set:
protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses.
Plant Journal 50, 347–363.
Kopka, J., Schauer, N., Krueger, S., Birkemeyer, C., Usadel, B., Bergmuller, E., Dormann, P.,
Weckwerth, W., Gibon, Y., Stitt, M., Willmitzer, L., Fernie, A.R. and Steinhauser, D. (2005)
GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics (Oxford, England) 21, 1635–
1638.
Kuromori, T., Hirayama, T., Kiyosue, Y., Takabe, H., Mizukado, S., Sakurai, T., Akiyama, K.,
Kamiya, A., Ito, T. and Shinozaki, K. (2004) A collection of 11,800 single-copy Ds transposon
insertion lines in Arabidopsis. Plant Journal 37, 897–905.
Kuromori, T., Wada, T., Kamiya, A., Yuguchi, M., Yokouchi, T., Imura, Y., Takabe, H., Sakurai, T.,
Akiyama, K., Hirayama, T., Okada, K. and Shinozaki, K. (2006) A trial of phenome analysis
using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant Journal 47, 640–
651.
Kusano, M., Fukushima, A., Arita, M., Jonsson, P., Moritz, T., Kobayashi, M., Hayashi, N., Tohge, T.
and Saito, K. (2007) Unbiased characterization of genotype-dependent metabolic regulations by
metabolomic approach in Arabidopsis thaliana. BMC Systems Biology 1, 53.
Last, R.L., Jones, A.D. and Shachar-Hill, Y. (2007) Towards the plant metabolome and beyond.
Nature Reviews Molecular Cell Biology 8, 167–174.
Li, S., Armstrong, C.M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P.O., Han, J.D.,
Chesneau, A., Hao, T., Goldberg, D.S., Li, N., Martinez, M., Rual, J.F., Lamesch, P., Xu, L.,
Tewari, M., Wong, S.L., Zhang, L.V., Berriz, G.F., Jacotot, L., Vaglio, P., Reboul, J., Hirozane-
Kishikawa, T., Li, Q., Gabel, H.W., Elewa, A., Baumgartner, B., Rose, D.J., Yu, H., Bosak, S.,
Sequerra, R., Fraser, A., Mango, S.E., Saxton, W.M., Strome, S., Van Den Heuvel, S., Piano, F.,
Vandenhaute, J., Sardet, C., Gerstein, M., Doucette-Stamm, L., Gunsalus, K.C., Harper, J.W.,
Cusick, M.E., Roth, F.P., Hill, D.E. and Vidal, M. (2004) A map of the interactome network of
the metazoan C. elegans. Science 303, 540–543.
Llave, C., Kasschau, K.D., Rector, M.A. and Carrington, J.C. (2002) Endogenous and silencing-
associated small RNAs in plants. The Plant Cell 14, 1605–1619.
Lu, C., Tej, S.S., Luo, S., Haudenschild, C.D., Meyers, B.C. and Green, P.J. (2005) Elucidation of the
small RNA component of the transcriptome. Science 309, 1567–1569.
Manfield, I.W., Jen, C.H., Pinney, J.W., Michalopoulos, I., Bradford, J.R., Gilmartin, P.M. and
Westhead, D.R. (2006) Arabidopsis Co-expression Tool (ACT): web server tools for microarray-
based gene expression analysis. Nucleic Acids Research 34, W504–509.
Matsuda, F., Yonekura-Sakakibara, K., Niida, R., Kuromori, T., Shinozaki, K. and Saito, K. (2009)
MS/MS spectral tag–based annotation of non-targeted profile of plant secondary metabolites,
Plant Journal 57, 555–577.
Miller, J.P., Lo, R.S., Ben-Hur, A., Desmarais, C., Stagljar, I., Noble, W.S. and Fields, S. (2005)
Large-scale identification of yeast integral membrane protein interactions. Proceedings of the
National Academy of Sciences of the United States of America 102, 12123–12128.
Analytical Platforms and Databases 241
Moco, S., Bino, R.J., Vorst, O., Verhoeven, H.A., de Groot, J., van Beek, T.A., Vervoort, J. and de
Vos, C.H. (2006) A liquid chromatography-mass spectrometry-based metabolome database for
tomato. Plant Physiology 141, 1205–1218.
Moco, S., Capanoglu, E., Tikunov, Y., Bino, R.J., Boyacioglu, D., Hall, R.D., Vervoort, J. and De
Vos, R.C. (2007) Tissue specialization at the metabolite level is perceived during the
development of tomato fruit. Journal of Experimental Botany 58, 4131–4146.
Mueller, L.A., Zhang, P. and Rhee, S.Y. (2003) AraCyc: a biochemical pathway database for
Arabidopsis. Plant Physiology 132, 453–460.
Nakano, M., Nobuta, K., Vemaraju, K., Tej, S.S., Skogen, J.W. and Meyers, B.C. (2006) Plant MPSS
databases: signature-based transcriptional resources for analyses of mRNA and small RNA,
Nucleic Acids Research 34, D731–735.
Nicholson, J.K., Connelly, J., Lindon, J.C. and Holmes, E. (2002) Metabonomics: a platform for
studying drug toxicity and gene function. Nature Reviews. Drug Discovery 1, 153–161.
Nobuta, K., Venu, R.C., Lu, C., Belo, A., Vemaraju, K., Kulkarni, K., Wang, W., Pillay, M., Green,
P.J., Wang, G.L. and Meyers, B.C. (2007) An expression atlas of rice mRNAs and small RNAs.
Nature Biotechnology 25, 473–477.
Obayashi, T., Kinoshita, K., Nakai, K., Shibaoka, M., Hayashi, S., Saeki, M., Shibata, D., Saito, K.
and Ohta, H. (2007) ATTED–II: a database of co–expressed genes and cis elements for
identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Research 35, D863–869.
Obayashi, T., Hayashi, S., Saeki, M., Ohta, H. and Kinoshita, K. (2009) ATTED-II provides
coexpressed gene networks for Arabidopsis. Nucleic Acids Research 37, D987–991.
Parkinson, H., Kapushesky, M., Shojatalab, M., Abeygunawardena, N., Coulson, R., Farne, A.,
Holloway, E., Kolesnykov, N., Lilja, P., Lukk, M., Mani, R., Rayner, T., Sharma, A., William, E.,
Sarkans, U. and Brazma, A. (2007) ArrayExpress–a public database of microarray experiments
and gene expression profiles. Nucleic Acids Research 35, D747–750.
Rajagopalan, R., Vaucheret, H., Trejo, J. and Bartel, D.P. (2006) A diverse and evolutionarily fluid
set of microRNAs in Arabidopsis thaliana. Genes & Development 20, 3407–3425.
Redman, J.C., Haas, B.J., Tanimoto, G. and Town, C.D. (2004) Development and evaluation of an
Arabidopsis whole genome Affymetrix probe array. Plant Journal 38, 545–561.
Riano-Pachon, D.M., Dreyer, I. and Mueller–Roeber, B. (2005) Orphan transcripts in Arabidopsis
thaliana: identification of several hundred previously unrecognized genes. Plant Journal 43, 205–
212.
Richly, E., Dietzmann, A., Biehl, A., Kurth, J., Laloi, C., Apel, K., Salamini, F. and Leister, D.
(2003) Covariations in the nuclear chloroplast transcriptome reveal a regulatory master-switch.
EMBO Reports
4, 491–498.
Robinette, S.L., Zhang, F., Bruschweiler-Li, L. and Bruschweiler, R. (2008) Web server based
complex mixture analysis by NMR. Analytical Chemistry 80, 3606–3611.
Rual, J.F., Venkatesan, K., Hao, T., Hirozane-Kishikawa, T., Dricot, A., Li, N., Berriz, G.F., Gibbons,
F.D., Dreze, M., Ayivi-Guedehoussou, N., Klitgord, N., Simon, C., Boxem, M., Milstein, S.,
Rosenberg, J., Goldberg, D.S., Zhang, L.V., Wong, S.L., Franklin, G., Li, S., Albala, J.S., Lim, J.,
Fraughton, C., Llamosas, E., Cevik, S., Bex, C., Lamesch, P., Sikorski, R.S., Vandenhaute, J.,
Zoghbi, H.Y., Smolyar, A., Bosak, S., Sequerra, R., Doucette-Stamm, L., Cusick, M.E., Hill,
D.E., Roth, F.P. and Vidal, M. (2005) Towards a proteome-scale map of the human protein-
protein interaction network. Nature 437, 1173–1178.
Saito, K., Hirai, M.Y. and Yonekura-Sakakibara, K. (2008) Decoding genes with coexpression
networks and metabolomics – 'majority report by precogs'. Trends in Plant Science 13, 36–43.
Sakurai, T., Satou, M., Akiyama, K., Iida, K., Seki, M., Kuromori, T., Ito, T., Konagaya, A., Toyoda,
T. and Shinozaki, K. (2005) RARGE: a large-scale database of RIKEN Arabidopsis resources
ranging from transcriptome to phenome. Nucleic Acids Research 33, D647–650.
Sawada, Y., Akiyama, K., Sakata, A., Kuwahara, A., Otsuki, H., Sakurai, T., Saito, K. and Hirai,
M.Y. (2009) Widely targeted metabolomics based on large-scale MS/MS data for elucidating
metabolite accumulation patterns in plants. Plant & Cell Physiology 50, 37–47.
242 Analytical Platforms and Databases
Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel,
D. and Lohmann, J.U. (2005) A gene expression map of Arabidopsis thaliana development,
Nature Genetics 37, 501–506.
Seki, M., Narusaka, M., Kamiya, A., Ishida, J., Satou, M., Sakurai, T., Nakajima, M., Enju, A.,
Akiyama, K., Oono, Y., Muramatsu, M., Hayashizaki, Y., Kawai, J., Carninci, P., Itoh, M., Ishii,
Y., Arakawa, T., Shibata, K., Shinagawa, A. and Shinozaki, K. (2002) Functional annotation of a
full-length Arabidopsis cDNA collection. Science 296, 141–145.
Shinbo, Y., Nakamura, Y., Altaf-Ul-Amin, M., Asahi, H., Kurokawa, K., Arita, M., Saito, K., Ohta,
D., Shibata, D. and Kanaya, S. (2006) KNApSAcK: A comprehensive species-metabolite
relationship database. Biotechnology in Agriculture and Forestry 57, 165–184.
Shoemaker, B.A. and Panchenko, A.R. (2007) Deciphering protein-protein interactions. Part I.
Experimental techniques and databases. PLoS Computational Biology 3, e42.
Small, I., Peeters, N., Legeai, F. and Lurin, C. (2004) Predotar: A tool for rapidly screening
proteomes for N-terminal targeting sequences. Proteomics 4, 1581–1590.
Steinhauser, D., Usadel, B., Luedemann, A., Thimm, O. and Kopka, J. (2004) CSB.DB: a
comprehensive systems-biology database. Bioinformatics (Oxford, England) 20, 3647–3651.
Sumner, L.W., Mendes, P. and Dixon, R.A. (2003) Plant metabolomics: large-scale phytochemistry
in the functional genomics era. Phytochemistry 62, 817–836.
Swarbreck, D., Wilks, C., Lamesch, P., Berardini, T.Z., Garcia-Hernandez, M., Foerster, H., Li, D.,
Meyer, T., Muller, R., Ploetz, L., Radenbaugh, A., Singh, S., Swing, V., Tissier, C., Zhang, P.
and Huala, E. (2008) The Arabidopsis Information Resource (TAIR): gene structure and function
annotation. Nucleic Acids Research 36, D1009–1014.
Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L.A., Rhee,
S.Y. and Stitt, M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto
diagrams of metabolic pathways and other biological processes. Plant Journal 37, 914–939.
Tikunov, Y., Lommen, A., de Vos, C.H., Verhoeven, H.A., Bino, R.J., Hall, R.D. and Bovy, A.G.
(2005) A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of
tomato fruit volatiles. Plant Physiology 139, 1125–1137.
Tohge, T., Nishiyama, Y., Hirai, M.Y., Yano, M., Nakajima, J., Awazuhara, M., Inoue, E., Takahashi,
H., Goodenowe, D.B., Kitayama, M., Noji, M., Yamazaki, M. and Saito, K. (2005) Functional
genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-
expressing an MYB transcription factor. Plant Journal 42, 218–235.
Tokimatsu, T., Sakurai, N., Suzuki, H., Ohta, H., Nishitani, K., Koyama, T., Umezawa, T., Misawa,
N., Saito, K. and Shibata, D. (2005) KaPPA-view: a web-based analysis tool for integration of
transcript and metabolite data on plant metabolic pathway maps. Plant Physiology 138, 1289–
1300.
Toufighi, K., Brady, S.M., Austin, R., Ly, E. and Provart, N.J. (2005) The Botany Array Resource:
e–Northerns, Expression Angling, and promoter analyses. Plant Journal 43, 153–163.
Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan,
V., Srinivasan, M., Pochart, P., Qureshi-Emili, A., Li, Y., Godwin, B., Conover, D., Kalbfleisch,
T., Vijayadamodar, G., Yang, M., Johnston, M., Fields, S. and Rothberg, J.M. (2000) A
comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403,
623–627.
Urbanczyk-Wochniak, E. and Sumner, L.W. (2007) MedicCyc: a biochemical pathway database for
Medicago truncatula. Bioinformatics (Oxford, England) 23, 1418–1423.
Usadel, B., Nagel, A., Thimm, O., Redestig, H., Blaesing, O.E., Palacios-Rojas, N., Selbig, J.,
Hannemann, J., Piques, M.C., Steinhauser, D., Scheible, W.R., Gibon, Y., Morcuende, R., Weicht,
D., Meyer, S. and Stitt, M. (2005) Extension of the visualization tool MapMan to allow statistical
analysis of arrays, display of coresponding genes, and comparison with known responses. Plant
Physiology 138, 1195–1204.
Villas-Boas, S.G., Mas, S., Akesson, M., Smedsgaard, J. and Nielsen, J. (2005) Mass spectrometry in
metabolome analysis. Mass Spectrometry Reviews 24, 613–646.
Analytical Platforms and Databases 243
Wang, Q.Z., Wu, C.Y., Chen, T., Chen, X. and Zhao, X.M. (2006) Integrating metabolomics into a
systems biology framework to exploit metabolic complexity: strategies and applications in
microorganisms. Applied Microbiology and Biotechnology 70, 151–161.
Wang, Y., Xiao, J., Suzek, T.O., Zhang, J., Wang, J. and Bryant, S.H. (2009) PubChem: a public
information system for analyzing bioactivities of small molecules. Nucleic Acids Research
W623–W633.
Wishart, D.S., Knox, C., Guo, A.C., Eisner, R., Young, N., Gautam, B., Hau, D.D., Psychogios, N.,
Dong, E., Bouatra, S., Mandal, R., Sinelnikov, I., Xia, J., Jia, L., Cruz, J.A., Lim, E., Sobsey,
C.A., Shrivastava, S., Huang, P., Liu, P., Fang, L., Peng, J., Fradette, R., Cheng, D., Tzur, D.,
Clements, M., Lewis, A., De Souza, A., Zuniga, A., Dawe, M., Xiong, Y., Clive, D., Greiner, R.,
Nazyrova, A., Shaykhutdinov, R., Li, L., Vogel, H.J. and Forsythe, I. (2009) HMDB: a
knowledgebase for the human metabolome. Nucleic Acids Research 37, D603–610.
Xie, Z., Allen, E., Fahlgren, N., Calamar, A., Givan, S.A. and Carrington, J.C. (2005) Expression of
Arabidopsis MIRNA genes. Plant Physiology 138, 2145–2154.
Yamada, K., Lim, J., Dale, J.M., Chen, H.M., Shinn, P., Palm, C.J., Southwick, A.M., Wu, H.C., Kim,
C., Nguyen, M., Pham, P., Cheuk, R., Karlin–Newmann, G., Liu, S.X., Lam, B., Sakano, H., Wu,
T., Yu, G.X., Miranda, M., Quach, H.L., Tripp, M., Chang, C.H., Lee, J.M., Toriumi, M., Chan,
M.M.H., Tang, C.C., Onodera, C.S., Deng, J.M., Akiyama, K., Ansari, Y., Arakawa, T., Banh, J.,
Banno, F., Bowser, L., Brooks, S., Carninci, P., Chao, Q.M., Choy, N., Enju, A., Goldsmith, A.D.,
Gurjal, M., Hansen, N.F., Hayashizaki, Y., Johnson-Hopson, C., Hsuan, V.W., Iida, K., Karnes,
M., Khan, S., Koesema, E., Ishida, J., Jiang, P.X., Jones, T., Kawai, J., Kamiya, A., Meyers, C.,
Nakajima, M., Narusaka, M., Seki, M., Sakurai, T., Satou, M., Tamse, R., Vaysberg, M.,
Wallender, E.K., Wong, C., Yamamura, Y., Yuan, S.L., Shinozaki, K., Davis, R.W., Theologis, A.
and Ecker, J.R. (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome.
Science 302, 842–846.
