Shen S., Tuszynski J.A. Theory and Mathematical Methods for Bioformatics
Подождите немного. Документ загружается.

424 14 Semantic Analysis for Protein Primary Structure
2. In database Ω, different core words do not contain each other. They occur
once and only once in database Ω.
3. When the vector of a core word contracts, this core word changes into the
τth ranked key word and the τ th ranked core word, thereby generating
the homologous structure tree of the protein. The homologous structure
tree is significant for long core words.
Searching for the Core Words in a Database
Searching for core words in a database is similar to that in a sequence. Here the
database is considered a long sequence of joined proteins and each protein is
distinguished by list separators. Thus the definition of core words in a database
requires the absence of the list separators in the core words. Hence we denote
by
Ω = {c
1
,c
2
, ··· ,c
n
},c
i
∈ V
q+1
, (14.23)
the sequence generated by the database, where the list separator 0 is added
to set V
q
= {1, 2, ··· , 20}, giving V
q+1
. The search for the core words in
a database is implemented by the following recursive computation.
Step 14.2.1 Take a positive integer k
0
, which follows the conditions below:
1. Any vector b
(k
0
)
in V
(k
0
)
q
occurs at least twice in database Ω.Thatis,
n
Ω
(b
(k
0
)
) ≥ 2alwaysholds.
2. In V
(k
0
+1)
q
, there is a vector b
(k
0
+1)
that occurs in database Ω only
once. That is, n
Ω
(b
(k
0
+1)
)=1holds.
We call k
0
the original rank of the recursive computation.
Step 14.2.2 Take k = k
0
+1,k
0
+2, ··· , and compute recursively. Here
c
(k)
i
=(c
i
,c
i+1
, ··· ,c
i+k−1
) ,i=0, 1, ··· ,n− k +1. (14.24)
From this, integers in the set N
k
= {0, 1, 2, ··· ,n− k +1} are classified
into two classes by c
(k)
i
.Theyare:
1. We define
N
k,1
=
i ∈ N
k
: n
Ω
c
(k)
i
=1
,
which is the subscript set of c
(k)
i
which occurs only once in Ω.
2. We define
N
k,2
=
i ∈ N
k
: n
Ω
c
(k)
i
> 1
,
which is the subscript set of c
(k)
i
which occurs more than once in Ω.
Then set
Ω
k
=
c
(k)
i
,i∈ N
k,1
,i∈ N
k−1,2
(14.25)
will be the collection of the kth ranked core words in database Ω.
14.2 Permutation and Combination Methods 425
Step 14.2.3 The subscript set of set N
k,2
given in Step 14.2.2 can also be
classified. Here, we take N
k,2,i
1
,N
k,2,i
2
, ··· ,N
k,2,i
p
to be a group of sub-
sets of N
k,2
.WedenotethesetD
k
= {i
1
,i
2
, ··· ,i
p
}, which follows the
conditions below:
1. If i
j
= i
j
,thenc
(k)
i
j
= c
(k)
i
j
must hold.
2. If i
∈ N
k,2,i
j
,thenc
(k)
i
= c
(k)
i
j
must hold. Therefore, sets N
k,2,i
1
,
N
k,2,i
2
, ··· ,N
k,2,i
p
are disjoint with each other.
3.
-
p
j=1
N
k,2,i
j
= N
k,2
. Thus, sets N
k,2,i
1
,N
k,2,i
2
, ··· ,N
k,2,i
p
are a divi-
sion of set N
k,2
.
From the classification N
k,2,i
1
,N
k,2,i
2
, ··· ,N
k,2,i
p
of N
k,2
, the recursive
computation can be continued as follows:
4. With every subscript in set N
k,2
as a starting point, construct k +1
dimensional vectors. That is, take vectors c
(k+1)
i
, i ∈ N
k,2
and repeat
Step 14.2.2, to obtain two subsets N
k+1,1
and N
k+1,2
of set N
k,2
,whose
definitions are the same as that of N
k,1
and N
k,2
in Step 14.2.2, classifi-
cations 1 and 2, respectively. At this time, Ω
k+1
= {c
(k+1)
i
,i∈ N
k+1,1
}
is the collection of the (k + 1)th ranked core words in database Ω.
For the elements in N
k+1,2
, we can repeat the computation in Steps
14.2.2 and 14.2.3 to construct the set N
k+2,1
and N
k+2,2
. Recurring
in this way, the collection of the core words in database Ω can be
obtained.
5. In the previous computation (4), the comparison of vectors c
(k+1)
i
, i ∈
N
k,2
can only be implemented in the divided set N
k,2,i
1
,N
k,2,i
2
, ···,
N
k,2,i
p
of N
k,2
. Here, each N
k,2,i
j
is divided into two sets N
k+1,1,i
j
,
N
k+1,2,i
j
, where the vectors c
(k+1)
i
corresponding to subscript i in
N
k+1,1,i
j
occur only once in c
(k+1)
i
, i
∈ N
k,2,i
j
, while the vectors
c
(k+1)
i
corresponding to subscript i in N
k+1,2,i
j
occur more than once
in c
(k+1)
i
, i
∈ N
k,2,i
j
.
The amount of computation can be greatly reduced in this way, which makes
the complexity of searching and computation for all core words in database Ω
a linear function of |Ω|. We refer to the above algorithm as recursive compu-
tation for nonlinear complexity and core words of a database.
With the discussion on the nonlinear complexity of databases, key words
and core words, proteins can be classified and aligned. In this book, we will
not discuss this problem; instead we focus on some special problems in the
next section.
14.2.4 Applications of Combinatorial Analysis
Besides what was mentioned in Sect. 14.1 (the application of analysis on pro-
tein structure using information and statistics, and the application of relative
entropy density dual theory), there are many applications of protein structure
analysis using combinatorial analysis. We discuss these as follows.
426 14 Semantic Analysis for Protein Primary Structure
Run Analysis on Databases of Protein Primary Sequences
We see from Sect. 14.1 that, several amino acids, such as A, R, N, etc., are
of very high affinity. Thus we assume that, vectors consisting of single letters
may form long polypeptide chains. If we name vectors consisting of single
letters as runs, then analysis on these special polypeptide chains will then be
called run analysis.
Suppose c
(s,t)
is a local sequence of sequence C.Ifc
s
= c
s+1
= ···= c
t
=
c ∈ V
q
, c
(s,t)
is to be called a run vector and t − s + 1 is to be called the run
length of this vector. We do statistical analysis on these run vectors in the
database Ω, and some results are summarized as follows:
1. The differences between the maximum run lengths of different amino acids
are quite significant. In the Swiss-Prot database, the maximum run lengths
of different amino acids are shown in Table 14.10.
2. We see from the computation in the Swiss-Prot database that I and W
are two special amino acids. The affinity of peptide chains with double I is
neutral, k(I, I) = log
p(I,I)
p(I)p(I)
=0.0964, while the affinity of peptide chains
with triple I is repulsive, k(I, I, I) = log
p(I,I,I)
p(I,I)p(I)
= −0.0926. The affinity of
peptide chains with double W or triple W is attractive, while the affinity
of peptide chains with W of four runs is repulsive. Here
⎧
⎪
⎪
⎪
⎪
⎪
⎪
⎪
⎨
⎪
⎪
⎪
⎪
⎪
⎪
⎪
⎩
k(W, W) = log
p(W, W)
p(W)p(W)
=0.5663 ,
k(W, W, W) = log
p(W, W, W)
p(W, W)p(W)
=0.5075 ,
k(W, W, W, W) = log
p(W, W, W, W)
p(W, W, W)p(W)
= −0.8316 .
3. Amino acids with long runs are often of high ranked Markovity. Here
k(c
1
,c
2
, ··· ,c
k+2
) − k(c
1
,c
2
, ··· ,c
k+1
)
=log
p(c
1
,c
2
, ··· ,c
k+2
)
p(c
1
,c
2
, ··· ,c
k
)
− log
p(c
1
,c
2
, ··· ,c
k+1
)
p(c
1
,c
2
, ··· ,c
k
)
=log
p(c
1
,c
2
, ··· ,c
k
)p(c
1
,c
2
, ··· ,c
k+2
)
p(c
1
,c
2
, ··· ,c
k+1
)p(c
1
,c
2
, ··· ,c
k
)
=log
p(c
1
,c
k+2
|c
2
, ··· ,c
k+1
)
p(c
1
|c
2
, ··· ,c
k+1
)p(c
k+2
|c
2
, ··· ,c
k+1
)
∼ 0 .
Table 14.10. The maximum run lengths of different amino acids
One-letter code A R N D C Q E G H I
Maximum run length 21 14 50 44 11 40 31 24 14 7
One-letter code L K M F P S T W Y V
Maximumrunlength19107 105042184 128

14.2 Permutation and Combination Methods 427
4. We see from the computation in the Swiss-Prot database that, for run
sequences of different amino acids, when the run lengths reach a certain
number, their transferences have a great propensity for orientation. For
example, when the run length of amino acid A reaches 12, the possibility
of its state transferring to A, G, S, V is very high, higher than 85%. This
characteristic is similar to the English word structure. For instance, in
English, what follows “qu” will always be one of the four letters a, e, i, o.
5. We can see from the fact that different amino acids have relatively long
runs, that the lexical structure of protein primary structure is quite differ-
ent from that of English and Chinese. In English or Chinese vocabularies,
the maximum run of single letters are often only 2, while that of single
letters in protein primary structure sequences may reach 50 (e.g., amino
acid P).
Search and Alignment of Homologous Proteins
We have stated in the above text that, if b
(k)
is a core word, it occurs in the
database Ω once, and only once. If vector b
(k)
is contracted back and forth,
then key words of different ranks can be formed in database Ω. Proteins with
these key words consist of the same peptide chains in considerably longer seg-
ments, where the corresponding homologous proteins and their stable regions
can be found.
Example 32. In Swiss-Prot version 2000, we have a core word with length 29:
GREFSLRRGDRLLLFPFLSPQKDPEIYTE .
The protein it locates and its starting site are CAML-MOUSE, 373. Its seg-
ments with length 7 and the serial number and sites of the segments occurring
in other proteins are
1 Segment Serial no. 2 Serial no. 2 Serial no. 2 Serial no. 2
4 RGDRLLL CAMG-MOUSE 379 CAML-HOMAN 379 CAML-MOUSE 380 CAML-RAT 380
4 GDRLLLF CAMG-MOUSE 380 CAML-HOMAN 380 CAML-MOUSE 381 CAML-RAT 381
4 LLLFPFL CAMG-MOUSE 383 CAML-HOMAN 383 CAML-MOUSE 384 CAML-RAT 384
4 LLFPFLS CAMG-MOUSE 384 CAML-HOMAN 384 CAML-MOUSE 385 CAML-RAT 385
4 LFPFLSP CAMG-MOUSE 385 CAML-HOMAN 385 CAML-MOUSE 386 CAML-RAT 386
3 FSLRRGD CAMG-MOUSE 375 CAML-MOUSE 376 CAML-RAT 376
3 PQKDPEI CAMG-MOUSE 391 CAML-MOUSE 392 CAML-RAT 392
3 SLRRGDR CAMG-MOUSE 376 CAML-MOUSE 377 CAML-RAT 377
Therefore, “1” is frequency number and “2” is location. The vector segment
GREFSLRRGDRLLLFPFLSPQKDPEIYTE is a marker of the homology of
gene CAML-MOUSE, CAMG-MOUSE, and CAML-RAT. We can also search
for several core words in the same protein, and thereby obtain its homologous
proteins.
428 14 Semantic Analysis for Protein Primary Structure
The Cutting of Protein Sequences and the Prediction
of Homologous Proteins
We analyze the cutting of protein sequences and the prediction of homologous
proteins by using protein RISA-CHLPN and RISA-CHLTR in Example 30. We
denote these two protein sequences as C and D respectively. Their nonlinear
complexity and nonsingular complexity are both 4. Thus, the fourth ranked
DG graph G
4
(C),G
4
(D) generated by C and D are antitrunk trees with two
branches. We can then discuss the cutting of these proteins:
1. We compare the primary structure of proteins C and D given in Exam-
ple 30. The lengths of these two sequences are both n = 289, where the
amino acids differ in ten sites and stay the same in the remaining sites.
The differing sites are 7, 18, 50, 63, 94, 123, 139, 234, 272, and 280. We
call these cutting sites.
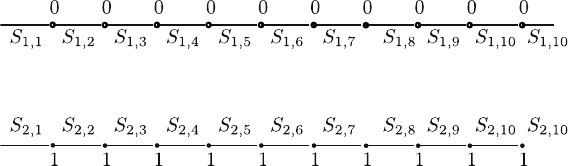
2. As is shown in Fig. 14.2, we draw proteins C and D into two parallel lines.
In these parallel lines, the lines present the same amino acids, while the
sites corresponding to the hollow points and solid points present differ-
ent amino acids. If we hold the lines presenting the same amino acids
and randomly choose amino acids from sequence C or D to the sites
where the amino acids are different in C and D, this operation is called
the cutting operation of homologous proteins. This operation actually di-
vides two homologous proteins into several segments and then combines
them in the original order. If the original proteins C and D are denoted
by
C = S
1,1
0S
1,2
0S
1,3
0S
1,4
0S
1,5
0S
1,6
0S
1,7
0S
1,8
0S
1,9
0S
1,10
0S
1,11
,
and
D = S
2,1
1S
2,2
1S
2,3
1S
2,4
1S
2,5
1S
2,6
1S
2,7
1S
2,8
1S
2,9
1S
2,10
1S
2,11
,
then the cutting sequence generated by this is
C
= S
1,1
0S
1,2
1S
2,3
1S
2,4
1S
2,5
0S
1,6
0S
1,7
0S
1,8
1S
2,9
1S
2,10
0S
1,11
.
Fig. 14.2. Primary structure relationship between protein RISA-CHLPN and pro-
tein RISA-CHLTR
14.3 Exercises, Analyses, and Computation 429
3. We reach the following conclusion: If there are k cutting points in two
proteins, then there would be 2
k
different combinations for this cutting
procedure. All of these combinations of cutting may generate new homol-
ogous proteins.
From this we can predict that, there may be 2
10
∼ 1000 kinds of homologous
trichosanthin structures. Biological experimentation is required to demon-
strate and illustrate whether these homologous proteins exist and what their
function and characteristics are. One particularly significant role of bioin-
formaticsistoprovidethescope,content and direction of experiments for
biologists, thereby greatly decreasing the number of the possible experiments.
The cutting of sequences provides a tool for this.
Semantic analysis for biological sequences is an important problem that
combines mathematics and biology. We see from the above discussion that
the corresponding analysis is related to the essential problems of the life sci-
ences. Thus, a close relationship exists between the life sciences and biological
engineering. Both the depth and breadth of the discussions in this book are
preliminary. If the theories and methods in biology, mathematics and biologi-
cal computation are further combined and different types of databases, such as
databases of protein three-dimensional structures, protein functions, special
(such as enzyme, antibody, virus, etc.) databases, GenBank, cDNA database,
genome database are synthetically analyzed, its development and applications
will lead to significant progress in biology and the related sciences. These in-
clude the core problems in genometics, proteomics theory, the applications of
biological engineering and drug design, etc. We hope this book can stimulate
further research using a synergistic combination of mathematics and biology.
14.3 Exercises, Analyses, and Computation
Exercise 68. Explain the biological significance of “local words,” “key words,”
and “core words.”
Exercise 69. Explain the meaning of “relative entropy density function” in
protein primary structure.
Exercise 70. Perform the following computation for bacteria or archaebac-
teria (see the data given in [99]):
1. Take k = 9. Compute the probability distribution p(b
(9)
)ofb
(9)
∈ V
(9)
4
,
and the marginal distribution p
1
(b
(3)
,p
0
(b
1
)).

430 14 Semantic Analysis for Protein Primary Structure
2. For
q
0
b
(9)
= p
0
(b
1
)p
0
(b
2
)p
0
(b
3
)p
0
(b
4
)p
0
(b
5
)p(b
6
)p
0
(b
7
)p
0
(b
8
)p(b
0
) ,
q
1
b
(9)
= p
0
(b
1
,b
2
,b
3
)p
0
(b
4
,b
5
,b
6
)p
0
(b
7
,b
8
,b
0
) ,
q
2
b
(9)
= p
0
(b
4
,b
5
,b
6
)p
0
(b
7
,b
8
,b
9
)p
0
(b
1
,b
2
,b
3
) ,
q
3
b
(9)
= p
0
(b
7
,b
8
,b
9
)p
0
(b
1
,b
2
,b
3
)p
0
(b
4
,b
5
,b
6
) ,
compute the relative entropy density functions
k
s
b
(9)
=log
2
p
b
(9)
q
s
b
(9)
,s=0, 1, 2, 3 ,
and give the table of their relative entropy density functions.
3. Based on task 2, compute the mean, variance, and standard deviation of
k
s
(b
(9)
).
4. For τ =2.5, find the “local words” generated by k
s
(b
(9)
).
Exercise 71. Find the “key words” and “core words” generated by these
bacteria or archaebacteria.

Epilogue
In this book, we introduced the stochastic models for DNA (or RNA) mu-
tations, the theory of modulus structure used for gene recombination, gene
mutation and gene alignment, the fast alignment algorithms for pairwise and
multiple sequences, and the topological and graphical structures on the set
of outputs induced from multiple alignments, respectively. These new con-
cepts illustrate the large number of approaches for analyzing the structures
of biosequences, many of which strongly rely on mathematics. We hope that
the theory of the modulus structure will lead to new advances in algebra the-
ory and will play an important role in the study of gene recombination and
mutation.
On the other hand, this book is only the first step in what we expect will
become a long history of applying mathematics to improve both the depth
and meaning within the study of biology. Rather than developing the math-
ematical theory and methods in a vacuum, it is far more interesting when the
mathematics evolves in order to solve problems in biology, biomedicine, and
biomedical engineering.
Currently, we are in an ideal period for advancement in the life sciences,
and many scientists from disciplines other than biology are facing the chal-
lenge of working in this field. Incorporating mathematics into the life sciences
can only enhance the quality and accuracy of research in the life sciences. The
question of how to ideally incorporate these two disciplines is of great im-
portance, and it is our hope that this book will contribute towards this goal.
Although every attempt was made to ensure correctness and accuracy in this
edition before publication, the authors welcome comments and suggestions so
that any remaining errors or omissions may be addressed in future editions.

References
1. Allen, F.H. The Cambridge Structural Database: a quarter of a million crystal
structures and rising, Acta Crystallogr. Sect. B 58, 380–388 (2002)
2. Altekar, G., Dwarkadas, S., Huelsenbeck, J.P., and Ronquist, F. Parallel
Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic in-
ference, Bioinformatics 20, 407–415 (2004)
3. Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. Basic
local alignment search tool, J. Mol. Biol. 215, 403–410 (1990)
4. Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller,
W., and Lipman, J.D. Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs, Nucleic Acids Res. 25, 3389–3402 (1997)
5. Andreeva, A., Howorth, D., Brenner, S.E., Hubbard, T.J.P., Chothia, C.,
Murzin, A.G. SCOP database in 2004: refinements integrate structure and
sequence family data, Nucleic Acids Res. 32, 226–229 (2004)
6. Anfinsen, C.B. Principles that govern the folding of protein chains, Science
181:223–230 (1973)
7. Attwood, T.K. and Parry-Smith, D.J. Introduction to bioinformatics (Long-
man, Harlow, 1999)
8. Bairoch, A., Boeckmann, B., Ferro, S., and Gasteiger, E. Swiss-Prot: juggling
between evolution and stability, Brief. Bioinform. 5, 39–55 (2004)
9. Barker, W.C., Garavelli, J.S., Haft, D.H., Hunt, L.T., Marzec, C.R., Or-
cutt, B.C., Srinivasarao, G.Y., Yeh, L.S.L., Ledley, R.S., Mewes, H.W., Pfeif-
fer, F., and Tsugita, A. The PIR—international protein sequence database,
Nucleic Acids Res. 26, 27–32 (1998)
10. Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., and Wheeler, D.L.
GenBank, Nucleic Acids Res. 31, 23–27 (2003)
11. Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., and Wheeler, D.L.
GenBank: update, Nucleic Acids Res. 32, 23–26 (2004)
12. Berge, C. Hypergraphs–combinatorics of finite sets (North-Holland, Amster-
dam, 1989)
13. Berman, H.M., Westbrook, J., Feng, Z., and Bourne, P.E. The Protein Data
Bank, Nucleic Acids Res. 28, 235–242 (2000)
14. Chapman, M.S. Mapping the surface properties of macromolecules, Protein
Sci. 2, 459–469 (1993)
434 References
15. Chan, S.C., Wong, A.K.C., and Chiu, D.K.Y. A survey of multiple sequence
comparision methods, Bull. Math. Biol. 54(4), 563–598 (1992)
16. Chao, K.-M., Pearson, W.R., and Miller, W. Aligning two sequences within
a specified diagonal band, CABIOS 8, 481–487 (1992)
17. Chao, K.-M., Hardison, R.C., and Miller, W. Recent developments in linear-
space aligning methods: a survey, Comput. Appl. Biosci. 1(4), 271–291 (1994)
18. Chao, K.-M., Ostell, J., and Miller, W. A local aligning tool for very long
DNA sequences, Comput. Appl. Biosci. 11(2), 147–153 (1995)
19. Chao, K.-M., Zhang, J., Ostell, J., and Miller, W. A tool aligning very similar
DNA sequences, Comput. Appl. Biosci. 13(1), 75–80 (1997)
20. Chen, K.X., Jiang, H.L., and Ji, R.Y. Computer-aided drug design (Shanghai
Scientific & Technical, Shanghai, 2000)
21. Chew, L.P., Kedem, K., Huttenlocher, D.P., and Kleinberg, J. Fast detection
of geometric substructure in proteins, J. Comput. Biol. 6(3–4), 313–325 (1999)
22. CLUSTAL-W. http://www.ebi.ac.uk/Tools/clustalw/index.html (2007)
23. Cover, T.M. and Thomas, J.A. Elements of information theory (Wiley, New
York, 1991)
24. Dayhoff, M.O., Schwartz, R.M., Orcutt, B.C. A model of evolutionary change
in proteins. In: Dayhoff, M.O. (ed.) Atlas of protein sequence and structure,
vol. 5, suppl. 2, pp. 345–352 (National Biomedical Research Foundation, Wash-
ington, 1978)
25. Doob, J.L. Stochastic processes (Wiley, New York, 1953)
26. Eisenberg, D., Schwarz, E., Komaromy, M., and Wall, R. Analysis of mem-
brane and surface protein sequences with the hydrophobic moment plot,
J. Mol. Biol. 179, 125–142 (1984)
27. Elshorst, B., Hennig, M., Forsterling, H., Diener, A., Maurer, M. et al. NMR
solution structure of a complex of calmodulin with a binding peptide of the
Ca
2+
pump, Biochemistry 38(38), 12320–12332 (1999)
28. Felsenstein, J. Evolutionary trees from DNA sequences: a maximum likelihood
approach, J. Mol. Evol. 17, 368–376 (1981)
29. Felsenstein, J. PHYLIP (the PHYLogeny inference package), version 3.66
(University of Washington, Seattle, 2007)
30. Fitch, W.M. Toward defining the course of evolution: minimum change for
a specified tree topology, Syst. Zool. 20, 406–416 (1971)
31. Gallo, G., Longo, G., Nguyen, S., and Pallottino, S. Directed hypergraphs and
applications, Discrete Appl. Math. 42, 177–201 (1993)
32. Garavelli, J.S., Hou, Z., Pattabiraman, N., and Stephens, R.M. The RESID
database of protein structure modifications and the NRL-3D sequence-
structure database, Nucleic Acids Res. 29(1), 199–201 (2001)
33. Gasteiger, E., Jung, E., and Bairoch, A. SWISS-PROT: Connecting biological
knowledge via a protein database, Curr. Issues Mol. Biol. 3, 47–55 (2001)
34. Goldman, N. and Yang, Z. A codon-based model of nucleotide substitution
for protein-coding DNA sequences, Mol. Biol. Evol. 11, 725–735 (1994)
35. Golomb, S.W. Shift register sequences (Holden-Day, San Francisco, 1967)
36. Goonet, G.H., Korostensky, C., and Benner, S. Evaluation measures of multi-
ple sequence alignments, J. Comput. Biol. 7, 261–276 (2000)
37. Green, P.J. Reversible jump Markov chain Monte Carlo computation and
Bayesian model determination, Biometrika 82, 711–732 (1995)
38. Hasegawa, M., Kishino, H., and Yano, T. Dating of the human-ape splitting
by a molecular clock of mitochondrial DNA, J. Mol. Evol. 22, 160–174 (1985)
