Moss Tom. DNA-protein interactions: principles and protocols
Подождите немного. Документ загружается.


Genetic Analysis of DNA-Binding Proteins 431
431
From:
Methods in Molecular Biology, vol. 148: DNA–Protein Interactions: Principles and Protocols, 2nd ed.
Edited by: T. Moss © Humana Press Inc., Totowa, NJ
30
Genetic Analysis of DNA–Protein Interactions
Using a Reporter Gene Assay in Yeast
David R. Setzer, Deborah B. Schulman,
and Michael J. Bumbulis
1. Introduction
Understanding the underlying structural and physico-chemical basis for the
recognition of specific DNA sequences by regulatory proteins is a central goal
of modern biochemical genetics. A method for the rapid identification of
mutant molecules altered in the affinity and/or specificity of such interactions
could be a powerful tool in the hands of those studying this difficult problem.
Conventional genetic approaches for obtaining and analyzing interesting
mutant forms of specific DNA-binding proteins are often infeasible because of
the genetic intractability of the species being studied or as a result of difficul-
ties in identifying relevant and specific phenotypes associated with alterations
in the interaction under investigation. High-resolution genetic analysis of
DNA–protein interactions is particularly problematic in metazoans. We have
devised an approach that makes use of the modularity in structure and function
of eukaryotic transcription factors (1), the power of the polymerase chain reac-
tion (PCR) to generate specific DNA fragments with defined levels of
mutagenesis in vitro (2,3), and the recombinogenic potential of S. cerevisiae
(2,4) to carry out a high-resolution genetic analysis of the sequence-specific
DNA-binding properties of Xenopus transcription factor IIIA (TFIIIA) (5). It
seems likely that this approach will be generally applicable to the study of
many DNA–protein interactions.
1.1. Outline of the Approach
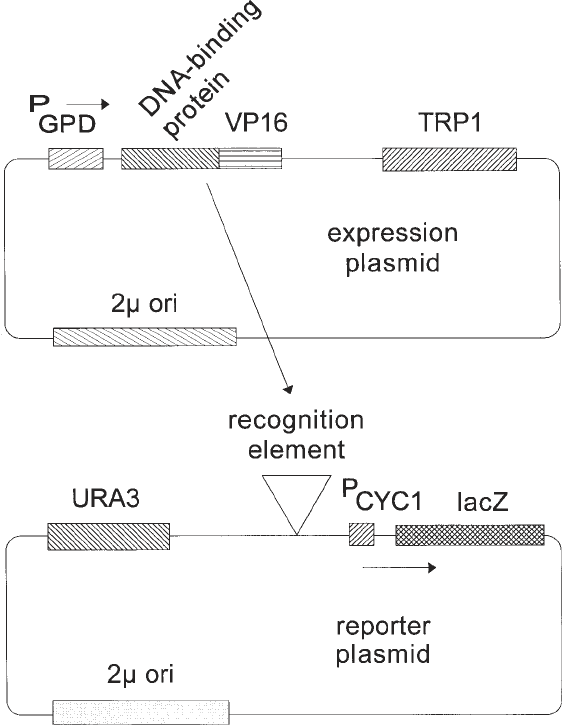
In its simplest form, the method we describe here involves the construction
and introduction of two plasmids into an appropriate yeast strain (Fig. 1). The
432 Setzer, Schulman, and Bumbulis
first, called the reporter plasmid, contains a reporter gene (Escherichia coli
β-galactosidase) under control of the core promoter of the S. cerevisiae iso-
1-cytochrome-c (CYC1) gene. In the parent plasmid, the upstream activator
sequence (UAS) normally required for expression from the CYC1 promoter
has been deleted so that β-galactosidase is expressed at very low levels, and
yeast strains containing this plasmid are white on X-gal indicator plates. A
DNA fragment containing the cognate recognition site for the DNA-binding
Fig. 1. Schematic representation of generic expression and reporter plasmids
derived from pG1 and p∆SS, respectively. Only the plasmid components functional in
yeast cells are shown; the plasmids also contain colE1 origins and β-lactamase genes
for replication and selection in E. coli.
Genetic Analysis of DNA-Binding Proteins 433
protein to be analyzed is substituted for the normal UAS. In the second plas-
mid, called the expression construct, the VP16 activation domain from Herpes
simplex is fused in-frame to a sequence encoding the DNA-binding domain of
the protein of interest. The DNA-binding-domain–VP16 fusion protein is
expressed under control of the constitutive glyceraldehyde-3-phosphate dehy-
drogenase (GPD) promoter of S. cerevisiae. The reporter and expression plas-
mids carry different selectable markers (URA3 and TRP1, for example) so that
both can be selected and maintained in an appropriate yeast strain (ura3
–
trp1
–
,
for example). When both plasmids are introduced into a single yeast cell and if
the DNA-binding domain of the protein of interest binds with sufficiently high
affinity and specificity to its recognition site in the reporter construct, the
VP16 activation domain will be displayed in the vicinity of the core CYC1
promoter and result in activation of transcription of the β-galactosidase
reporter gene. On X-gal indicator plates, such a strain will be blue. Thus, this
blue phenotype can be used as a marker for high-affinity interaction of the
DNA-binding domain of interest with its recognition sequence. Mutations in
either the DNA-binding protein or the DNA sequence to which it binds may
adversely affect binding, resulting in white or light blue colonies, or may
increase the affinity of binding, resulting in dark blue colonies (see Notes 1–5).
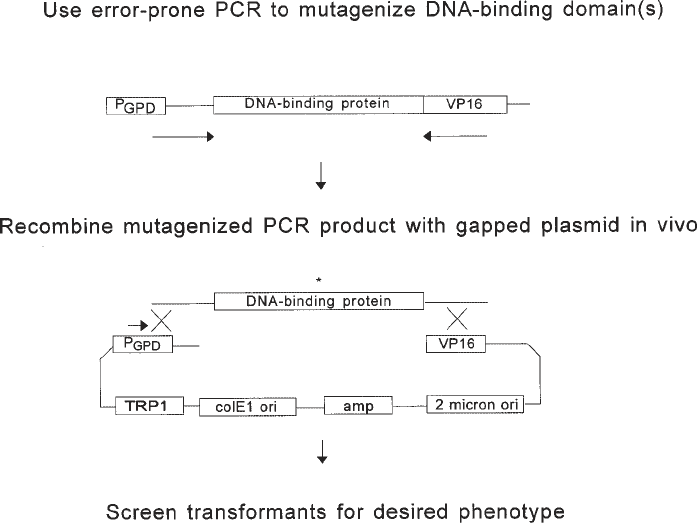
Generation of randomly mutated sequences encoding either the protein of
interest or its cognate recognition site and the introduction of these mutated
sequences into their appropriate contexts in either the expression or reporter
plasmids is achieved with technical ease and high efficiency using a combina-
tion of error-prone PCR in vitro and homologous recombination in vivo fol-
lowing transformation (Fig. 2). Unusually long oligonucleotide primers are
used for error-prone PCR (about 60–70 nucleotides, but see Note 6). The 3'
portion of these primers anneals to a substrate plasmid at sites flanking the
sequence to be mutagenized. The primers also contain 5' sequences identi-
cal to those which flank the ends of a linear version of the plasmid construct
into which the mutated sequences are to be introduced. Error-prone PCR is
used to synthesize a population of mutant DNA fragments containing the
sequence of interest flanked by the sequences defined by the long amplifica-
tion primers. In parallel, the plasmid into which the mutagenized PCR product
is to be inserted is linearized or gapped by digestion at one or two sites, respec-
tively, such that the unique ends of the PCR product correspond to sequences
at the ends of the linear or gapped plasmid. When cotransformed into compe-
tent yeast cells, the linear plasmid and mutant PCR products undergo homolo-
gous recombination in vivo to produce a circular plasmid product in which the
mutagenized fragment is integrated into the target plasmid at or between the
restriction sites used in linearization/gapping. Successful recombination events
can be clonally selected using the marker (URA3 or TRP1, for example) on the
434 Setzer, Schulman, and Bumbulis
target plasmid. If the yeast strain used for transformation already contains the
second plasmid component of the system, then resulting colonies containing
both reporter and expression plasmids can be selected and screened subse-
quently by replica-plating on indicator plates to identify mutants resulting in
altered phenotypes (white, light blue, or dark blue). After further tests to
ensure the mutant phenotype is authentic and the mutant protein or DNA
sequence is likely to be of interest, the mutant plasmid is recovered in E. coli
and the mutation identified by DNA sequence analysis, If desired, the mutant
DNA–protein interaction can be subjected to detailed biochemical or further
genetic analysis.
Fig. 2. Error-prone PCR in vitro and homologous recombination in vivo to
mutagenize the DNA-binding protein of interest and introduce the mutagenized frag-
ment into the expression vector. The asterisk represents a mutation introduced during
error-prone PCR. In the example shown here, the substrate used for error-prone PCR
is the expression plasmid itself, derived in this case from pG1, and the entire DNA-
binding protein is subjected to mutagenesis. It is also possible to target only a portion
of the DNA-binding protein for mutagenesis or to use a different plasmid as a sub-
strate for PCR, provided the expression plasmid and amplification primers are appro-
priately designed.
Genetic Analysis of DNA-Binding Proteins 435
2. Materials
2.1. Initial Design and Testing of the System
1. S. cerevisiae strain BJ2168 (6) or other haploid strain with appropriate genotype
(stable mutant alleles of genes used as selectable markers in expression and
reporter plasmids) (see Note 1).
2. p∆SS (7) or other plasmid to be used as parent for reporter plasmid construction
(see Note 2).
3. pG1 (7) or other plasmid to be used as parent for expression plasmid construction
(see Note 3).
4. pSJT-1193-CRF1 (8) or other source of DNA encoding the VP16 activation
domain (see Note 4)
5. Source of DNA encoding the DNA-binding protein or DNA-binding domain
of interest.
6. Source of DNA including the recognition sequence for the DNA-binding protein
or DNA-binding domain of interest.
7. Complete medium (C) agar plates lacking appropriate nutrients to permit selec-
tion of yeast strains containing reporter and expression plasmids. These will
include C-uracil, C-tryptophan, and C-uracil–tryptophan for systems making use
of derivatives of p∆SS and pG1. Procedures for preparation of liquid C medium
and C agar are described by Rose et al. (9). Our specific procedures are as follows:
a. Dissolve in 1 L water the following: 20 g dextrose, 20 g Bactoagar, 1.7 g
yeast nitrogen base without amino acids and without ammonium sulfate, 5 g
ammonium sulfate, and 0.5 g amino acid mixture (see step 7b). Autoclave to
sterilize and use to pour approx forty 100-mm plates.
b. The amino acid mixture used to prepare C agar plates contains 0.2 g arginine,
0.2 g histidine, 0.5 g lysine, 0.4 g methionine, 0.2 g phenylalanine, 0.4 g
tryptophan, 2.0 g threonine, 0.4 g tyrosine, 0.5 g serine, 0.2 g adenine, and 0.1 g
uracil. For selective plates, the appropriate combination of nutrients (uracil
and tryptophan [e.g., to select for plasmids containing URA3 and TRP1 mark-
ers]) should be omitted from the mixture.
8. SSX agar plates lacking appropriate nutrients as described in step 7, but also
containing 40 µg/mL X-gal, prepared as follows:
a. Dissolve the following in 900 mL water: 1.7 g yeast nitrogen base without
amino acids and without ammonium sulfate, and 5 g ammonium sulfate, 20 g
dextrose, 14 g Sigma agar, 0.5 g appropriate amino acid mixture (see step
7b). Autoclave to sterilize.
b. Cool to 48°C and add aseptically: 1 mL of 40 mg/mL X-gal prepared in N,N-
dimethylformamide and 100 mL 10X phosphate buffer (see step 8c).
c. 10X phosphate buffer is prepared by mixing the following in 1 L water: 136.1 g
KH
2
PO
4
(1 M), 19.8 g (NH
4
)
2
SO
4
(0.15 M), and 42.1 g KOH (0.75 N). Adjust
the pH to 7.0 and autoclave to sterilize.
9. Standard reagents and methods for subcloning DNA fragments into plasmids.
436 Setzer, Schulman, and Bumbulis
2.2. Error-Prone PCR
1. Plasmid(s) or other source(s) of DNA containing the sequence encoding the pro-
tein of interest and/or the DNA sequence recognized by the protein of interest.
2. Oligonucleotide primers that contain, at their 5' ends, approx 50 nucleotides of
sequence identity to the site immediately adjacent to the end of the linear plasmid
into which the PCR product is to be inserted. The 3' 15–20 nucleotides of
these primers should have sequence identity with the parts of the substrate
plasmid that define the DNA sequence to be amplified and mutagenized (see
Fig. 2 and Note 6).
2. Taq DNA polymerase.
3. Stock solutions of 100 mM MgCl
2
and 100 mM MnCl
2
.
4. 10X stock solution of Taq PCR buffer, lacking MgCl
2
: 100 mM Tris, pH 9.0,
500 mM KCl, and 1% Triton X-100 (Promega).
5. Individual stock solutions of dATP, dGTP, dCTP, and dTTP, each at a concen-
tration of 10 mM. In addition, individual stock solutions of the same, each at a
concentration of 2 mM.
6. Thermal cycler.
2.3. Yeast Transformation and Homologous Recombination
1. S. cerevisiae strain BJ2168 or other appropriate strain (see Note 1).
2. If the DNA-binding protein is to be mutagenized, BJ2168 containing the reporter
plasmid and BJ2168 containing the parent of the reporter plasmid (p∆SS, for
example).
3. If the DNA recognition site is to be mutagenized, BJ2168 containing the expres-
sion plasmid and BJ2168 containing the parent of the expression plasmid (pG1,
for example).
4. Linearized or gapped plasmid to be used as the target for integration of the PCR-
generated DNA fragment.
5. Crude product of the error-prone PCR
6. C-agar plates lacking the relevant nutrients for selection and maintenance of both
reporter and expression plasmids (uracil and tryptophan for p∆SS- and pG1-
derived plasmids).
7. Sterile stock solution of 100 mM lithium acetate, 1 mM EDTA, 10 mM Tris–Cl,
pH 8.0.
8. Sonicated salmon sperm DNA of about 10,000 bp average length, denatured by
heating to 100°C for 5 min at a concentration of about 5–10 mg/mL. Commer-
cially available salmon sperm DNA should be extracted with phenol/chloroform
and precipitated prior to use.
9. 40% (w/v) Polyethylene glycol (PEG) (average molecular weight of 3350). To
prepare this solution, autoclave 2 g solid PEG in a sealable tube. The PEG will
melt during sterilization and resolidify at room temperature. Many sterile PEG
aliquots can be prepared simultaneously. On the day of use, add 3.5 mL sterile
solution from step 7 to one of these aliquots, heat to 65°C, and mix vigorously to
Genetic Analysis of DNA-Binding Proteins 437
dissolve (final concentration of PEG is 40%). This quantity of solution is suffi-
cient for approx 10 transformations.
10. Sterile SOS solution: 2 mL 2 M sorbitol, 1.3 mL YEPD medium (9), 0.26 mL 100 mM
CaCl
2
, 0.4 mL H
2
O. YEPD medium: per liter of water, add 10 g Bactoyeast
extract, 20 g Bactopeptone, and 20 g glucose.
2.4. Screening for Mutants
1. C-agar plates lacking both uracil and tryptophan, and containing 40 mg/mL X-gal.
2. Liquid C-trp or C-ura medium for selection of only one of the two plasmids in BJ2168.
3. C-agar plates lacking uracil and tryptophan individually, as well as plates
lacking both.
4. Standard E. coli strain for plasmid transformation, propagation, and isolation,
along with reagents for distinguishing reporter and expression plasmids by
restriction endonuclease analysis.
5. 2% Sodium dodecyl sulfate (SDS)
6. Acid-washed glass beads (0.45 mm in diameter, from Sigma) prepared by wash-
ing overnight in 3 N HCl and then rinsing repeatedly in water.
7. Reagents for protein concentration determination using the BCA method (Pierce).
8. If possible, antibodies to the DNA-binding protein of interest and/or the activa-
tion domain used in the construction of the expression plasmid; reagents for
Western blotting.
2.5. Analysis of Mutants
1. Z buffer for determination of β-galactosidase activity: 60 mM Na
2
HPO
4
, 40 mM
NaH
2
PO
4
, 10 mM KCl, 1 mM MgSO
4
, and 40 mM β-mercaptoethanol, pH 7.0.
2. Other reagents for determination of β-galactosidase activity: chloroform, 0.1%
SDS, 4 mg/mL o-nitrophenol-β-D-galactoside (ONPG) prepared in Z buffer, and
1 M Na
2
CO
3
.
3. Reagents for DNA sequence determination.
3. Methods
3.1. Initial Design and Testing of the System
Details of the construction of appropriate reporter and expression constructs
will depend on the specific features of the plasmids and clones to be used. It is
therefore impossible to describe a step-by-step protocol for use in every case,
but standard recombinant DNA methods should suffice for preparation of
the desired plasmids. We will briefly outline the steps necessary for construc-
tion and testing of reporter and expression constructs prepared in p∆SS and
pG1, respectively.
3.1.1. Construction of the Reporter Plasmid
It is necessary that a DNA fragment containing one or more copies of the
DNA sequence recognized by the protein of interest be subcloned upstream of
438 Setzer, Schulman, and Bumbulis
the CYC1 core promoter in p∆SS. The only unique restriction site in p∆SS that
is suitable for insertion of such a fragment is an XhoI site. A DNA fragment
with XhoI-compatible ends and containing one or more copies of the relevant
DNA sequence should be subcloned into the XhoI site of p∆SS. Most typically,
this fragment would be either a restriction fragment from another plasmid or a
PCR product digested to produce XhoI-compatible ends. It is possible, and
probably desirable, to obtain p∆SS derivatives with multiple inserts of the DNA
sequence of interest, and with single inserts in either orientation. The number
and orientation of insert fragments must be diagnosed by some means, typi-
cally including restriction endonuclease mapping using enzymes that cut asym-
metrically within the insert fragment to determine orientation, PCR with
primers flanking the insert site to determine number of inserts, or DNA
sequence analysis to determine either orientation or number of inserts if the
insert fragment is not too long. An alternative to subcloning the insert fragment
into the XhoI site of p∆SS is to use homologous recombination as described in
Subheading 3.3. to integrate a PCR-generated DNA fragment into XhoI-
digested p∆SS. In this case, the PCR fragment should be produced under high-
fidelity conditions; even so, we recommend sequencing of the inserted
fragment in the resulting plasmid to ensure that no mutations were introduced
during amplification. If homologous recombination is used to generate the
reporter plasmid, judicious choice of sequences in the long primers used for
PCR can be used to regenerate either or both of the XhoI sites at the end of the
insert, or even to introduce novel restriction endonuclease recognition ele-
ments. This may facilitate the introduction of multiple copies of the DNA-
binding site into p∆SS.
3.1.2. Construction of the Expression Plasmid
One or more DNA fragments encoding an in-frame fusion of the DNA-bind-
ing protein of interest and a transcriptional activation domain must be intro-
duced into pG1 downstream of the GPD promoter. The unique SalI site in pG1
is probably the most convenient site for doing this. Fusion of the DNA-binding
protein of interest and the VP16 activation domain can be done directly in pG1
or, perhaps more conveniently, in a smaller, simpler plasmid vector and then
subcloned as a unit into pG1. Subsequent mutagenesis (Subheading 3.2.) can
be more directly targeted to the DNA-binding protein rather than to the activa-
tion domain if a unique restriction site (more precisely, one that does not occur
elsewhere in the plasmid outside of the sequence encoding the DNA-binding
protein) can be engineered at the junction of the DNA-binding protein and the
activation domain. It is also important to note that some DNA-binding pro-
teins, and particularly transcriptional activator proteins acting through the RNA
polymerase II core machinery, may contain endogenous transcriptional activa-
Genetic Analysis of DNA-Binding Proteins 439
tion domains that will function in S. cerevisiae; in that event, transcriptional
activity in the absence of the VP16 activation domain may be observed. The
existence of an intrinsic activation domain in the protein of interest might
obviate the need to prepare a fusion construct, but one must be careful in the
subsequent analysis to distinguish mutations affecting DNA-binding affinity
from those affecting transcriptional activation directly. The VP16 activa-
tion domain coding sequence followed by a polyadenylation signal from the
H. simplex thymidine kinase gene can be excised on a KpnI–HindIII fragment
of approx 760 bp from the plasmid pSJT-1193-CRF1 (8). At the KpnI cleavage
site, the reading frame for fusion to VP16 is XXG-GTA-CCX, but other plas-
mids in which the VP16 reading frame is shifted relative to the KpnI cleavage
site have also been constructed (8). The KpnI–HindIII fragment from this fam-
ily of constructs is suitable for preparing a fusion protein in which the VP16
domain is at the C-terminus. Depending on what is known about the polarity of
DNA binding by the protein of interest, this may or may not be desirable. Con-
struction of N-terminal fusions may be preferable in some cases (see Note 7);
these can be made by taking advantage of one of a number of vectors intended
for the construction of libraries for use in two-hybrid screens (e.g., pACT-II,
pGAD-GH, and pB42-AD from Clontech). One must be cautious in the choice
of vector, however (see Note 3). In the case of Xenopus TFIIIA binding to the
Xenopus 5S rRNA gene, some of these vectors (including pGAD10) result in
very low levels of protein expression and no detectable transcription activa-
tion. Also, see Note 4 concerning choice of activation domains vis-a-vis sensi-
tivity of the genetic assay.
3.1.3. In Vivo Assay of the Reporter and Expression Constructs
For each reporter plasmid constructed and the parent vector (p∆SS), as con-
trol, two strains derived from BJ2168 should be prepared, one containing the
expression plasmid in addition to the reporter, and one containing the parent
plasmid from which the expression plasmid was derived (pG1) in combination
with the reporter. The different selectable markers on these two plasmids
(URA3 and TRP1) allow their simultaneous maintenance in BJ2168 (ura3
–
trp1
–
) by selecting for growth in medium lacking uracil and tryptophan. The
requisite strains should be constructed by sequentially transforming BJ2168
with the reporter and expression plasmids. Methods for transformation of
BJ2168 (or other yeast strains) are described in detail in Subheading 3.3. One
need only adjust the protocol to reflect the nutritional requirements of the strain
being transformed and the selectable marker on the plasmid being introduced.
Thus, the doubly transformed strain would be selected on C-ura-trp plates.
Colonies of strains containing both reporter and expression plasmids can be
replica plated, spotted, or streaked onto C-ura-trp plates containing 40 µg/mL
440 Setzer, Schulman, and Bumbulis
X-gal. Colony color is assessed at an empirically determined time after robust
colony growth has occurred. For analysis of the Xenopus TFIIIA–5S rRNA
gene interaction, this was done typically after 2–3 d of growth at 30°C and an
additional 2–3 d at room temperature. For the system to be exploited success-
fully, one must be able to distinguish reproducibly the color of strains contain-
ing both the expression and reporter plasmids from that of all the other control
strains (lacking either expression of the fusion protein containing the DNA-
binding domain[s] of interest or the cognate recognition site in the reporter
construct, or both). If this is not the case, it may be possible to correct the
problem by manipulation of parameters as described in Note 5. Of course, it is
also possible that the particular interaction being studied will not be amenable
to analysis with this method; among other reasons, this could result from a
low-affinity/specificity interaction or from the existence of endogenous yeast
factors that interact with the binding site introduced into the reporter plasmid,
resulting in high levels of transcriptional activity in the absence of the interac-
tion being targeted for study.
3.2. Error-Prone PCR
The DNA-binding protein or its recognition site can be subjected to random
mutagenesis using error-prone PCR. In the following protocol, we assume that
the DNA-binding protein is targeted for mutagenesis, but the procedure can be
adapted readily for mutagenesis of the recognition site.
1. Set up a 50-µL polymerase chain reaction mixture containing 10–50 ng of plas-
mid DNA containing the sequence encoding the region to be mutagenized. This
can be the expression plasmid itself (see Note 8) or another plasmid containing
the sequence of interest. In addition, add 5 mL of 10X PCR buffer lacking MgCl
2
,
long amplification primers (see Subheading 2.2., Fig. 2, and Note 6) to a final
concentration of 0.3 µM each, three deoxynucleoside triphosphates to a final con-
centration of 1 mM each, the fourth deoxynucleoside triphosphate to a final con-
centration of 0.2 mM, MgCl
2
to a final concentration of 3 mM, MnCl
2
to a final
concentration of 0.05 mM, and 1 unit Taq DNA polymerase (see Note 9).
2. Amplify using a thermal cycler for 25 cycles, with each cycle being 94°C for
1 min, 42°C for 2 min, and 72°C for 1 min. After 25 cycles, use a final extension
step of 72°C for 7 min (see Note 9).
3. Use the crude PCR product (without purification) in a yeast transformation with
linearized/gapped target plasmid as described in Subheading 3.3.
3.3. Yeast Transformation and Homologous Recombination
1. Prepare a stock of linearized or gapped target plasmid by digesting to completion
with one or two restriction endoncleases that result in ends corresponding to the
site at which integration of the mutagenized DNA fragment is to occur. As an
example, with an expression plasmid derived from pG1, this might be a double
