Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.


influences the
accuraqt
of translation:
translation
can
be
made more
or less accurate
by changing
the
struc-
ture
of
165
rRNA. The
combination
of
the effects
of the
Sl2
protein
and
streptomycin
on the
rRNA
structure
explains
the behavior
of differ-
ent
mutants in
S 12,
some
of which
even make
the ribosome
dependent
onthe
presence
of
strep-
tomycin for
correct
translation.
We
now know
from
the crystal
structure
of
the ribosome
that l65
rRNA
is in
a
position
ro
make contacts
with
aminoacyl-tRNA.
TWo bases
of l65 rRNA
can
contact
the minor
groove
of
the helix formed
by
pairing
between
the anti-
codon in
IRNA
with the first
two
bases
of the
codon in mRNA.
This
directly
stabilizes
the struc-
ture
when the
correct
codon-anticodon
con-
tacts are made
at the
first two
codon
positions,
but it does
not monitor
contacts
at the third
position.
The
stabilization
of correctly
paired
amino-
acyl-tRNA may
have two
effects.
By holding
the
aminoacyl-tRNA
in
the A site,
it
prevents
it from
escaping
before the
next stage
of
protein
syn-
thesis. The
conformational
change in
the rRNA
may help to
trigger the
next
stage of the
reac-
tion,
which is the hydrolysis
of GTP
by EF-Tu.
Part of
the
proofreading
effect is determined
by timing.
An aminoacyl-tRNA
in
the A site
may
in effect
be trapped if
the next
stage
of
pro-
tein synthesis
occurs
while it
is there.
Thus a
delay between
entry into
the
A site and
pep-
tidyl
transfer may
give
more
opportunity
for a
mismatched
aminoacyl-tRNA
to dissociate. Mis-
matched
aminoacyl-tRNA
dissociates
more rap-
idly than
correctly matched
aminoacyl-tRNA,
probably
by a factor
of
-5x.
Its chance
of escap-
ing is therefore
increased
when
the
peptide
transfer
step is
slowed.
The
specificity
of decoding
has
been
assumed to reside
with the ribosome
itself, but
some recent results
suggest
that
translation fac-
tors influence
the
process
at
both the P
site and
A
site. An indication
that
EF-Tu is
involved in
maintaining
the reading
frame
is
provided
by
mutants
of
the
factor
that suppress
frameshift-
ing.
This implies
that EF-Tu
does not
merely
bring aminoacyl-tRNA
to the A
site, but also is
involved in
positioning
the incoming
amino-
acyl-IRNR relative
to the
pepridyl-tRNA
in the
P site.
A
striking case in which
factors
influence
meaning is found
at initiation.
Mutation
of the
AUG initiation
codon
to UUG in
the
yeast gene
Il1S4
prevents
initiation.
Extragenic
suppressor
mutations
can
be
found
that
allow
protein
syn-
thesis
to be initiated at
the mutant
UUG codon.
TWo
of these suppressors
prove
to be in
genes
coding for
the u and
p
subunits of eIF2,
the fac-
tor that
binds Met-tRNA to the P site.
The muta-
tion in eIFp2 resides in a
part
of the
protein
that
is almost
certainly
involved in
binding
nucleic
acid. It
seems likely that its target
is
either the
initiation
sequence of
mRNA as such or the
base-paired
association between the
mRNA
codon and tRNAlMet anticodon. This suggests
that
eIF2
participates
in the discrimination of
initiation
codons as well as bringing the
initia-
tor IRNA to the P site.
The
cost of
protein
synthesis
in terms of
high-energy
bonds may be
increased
by
proof
-
reading
processes.
An important
question
in
calculating the cost of
protein
synthesis
is
the
stage at
which the decision
is taken on whether
to accept
a IRNA.
If
a decision
occurs immedi-
ately to release an aminoacyl-tRNA-EF-TUGTP
complex,
there is
little
extra
cost for rejecting the
large
number of incorrect tRNAs that are
likely
(statistically)
to enter
the A site before the cor-
rect 1RNA is recognized. If, however, GTP is
hydrolyzed
before the
mismatched aminoacyl-
IRNA dissociates, the cost will be
greater.
A mis-
matched
aminoacyl-tRNA
can be rejected either
before or after the cleavage of GTP,
although
we
do not know
yet
where on average
it is
rejected.
There is some evidence
that the use
of GTP in vivo is
greater
than the three
high-
energy bonds that are used
in adding every
(cor-
rect)
amino acid to the chain.
Recoding Changes
Codon
Meanings
.
Changes in codon meaning can
be caused by
mutant tRNAs or by tRNAs
with special
properLies.
.
The reading frame can be changed by
frameshifting or bypassing, both
of which
depend
on
properties
of the
mRNA.
The reading frame of a
messenger usually is
invariant.
Tlanslation starts
at an AUG codon
and continues in triplets to a
temination codon.
Reading
takes
no notice of sense:
insertion or
deletion of a base causes
a frameshift
mutation,
in
which
the reading frame
is changed beyond
the
site of
mutation. Ribosomes and
tRNAs con-
tinue ineluctably in triplets, synthesizing
an
entirely different series of amino
acids.
There are
some
exceptions
to the usual
pat-
tern
of
translation that enable
a reading frame
with an interruption of
some sort-such
as a
9.16 Recoding Changes Codon
Meanings ztt
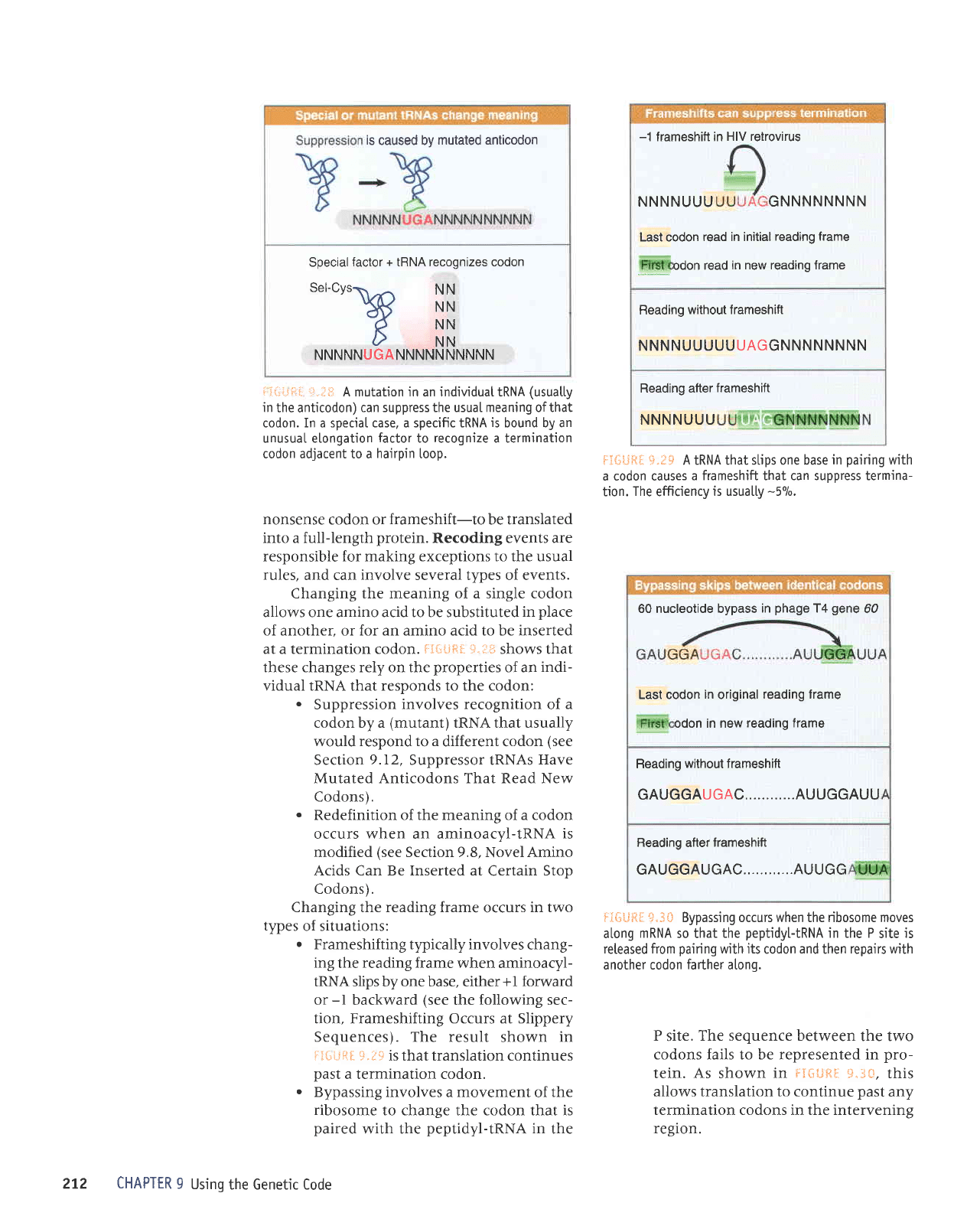
is
caused by
mutated anticodon
Special
factor
+
IRNA recognizes codon
s+cvs\sg
Nil
8ilil
NNNNNUGANNNNNNNNNN
a:{i-jei
'-:.:l:l
A mutation in an individuat tRNA
(usuatty
in
the anticodon)
can suppress the usuaI meaning ofthat
codon. In a spec'iaI case, a specific IRNA
is
bound by an
unusual elongation
factor
to
recognize
a termination
codon adjacent to a
hairpin
[oop.
nonsense
codon or
frameshift-to
be translated
into a full-length
protein.
Recoding events are
responsible for making exceptions to the usual
rules,
and can involve several types of events.
Changing the meaning of a single codon
allows one amino acid to be substituted
in
place
of another, or for an amino acid to be inserted
at a termination codon.
f.i*tiFtE *"t:s
shows that
these changes rely on the
properties
of an
indi-
vidual IRNA
that
responds to the codon:
.
Suppression
involves recognition
of
a
codon by a
(mutant)
IRNA that usually
would respond to a
different codon
(see
Section
9.I2,
Suppressor tRNAs
Have
Mutated Anticodons That Read New
Codons).
.
Redefinition of the meaning of a codon
occurs when an aminoacyl-tRNA
is
modified
(see
Section 9.8,
Novel
Amino
Acids Can Be Inserted at
Certain
Stop
Codons).
Changing the reading frame
occurs
in two
types
of situations:
.
Frameshifting typically involves
chang-
ing
the
reading frame
when aminoacyl-
IRNA slips by one base, either
+l
forward
or
-l
backward
(see
the following sec-
tion, Frameshifting Occurs at Slippery
Sequences). The result
shown
in
a:*i,jt[
*.iF is that translation continues
past
a termination codon.
.
Bypassing involves
a
movement
of
the
ribosome
to change the codon that is
paired
with the
peptidyl-tRNA
in
the
Usinq
the Genetic Code
-1
frameshift in
HIV retrovirus
NNNNUUU GNNNNNNNN
Last codon
read in initial
reading frame
odon read
in new reading lrame
Reading
without f rameshift
NNNNUUUUUUAGGNNNNNNNN
Reading after
frameshift
NNNNUUU
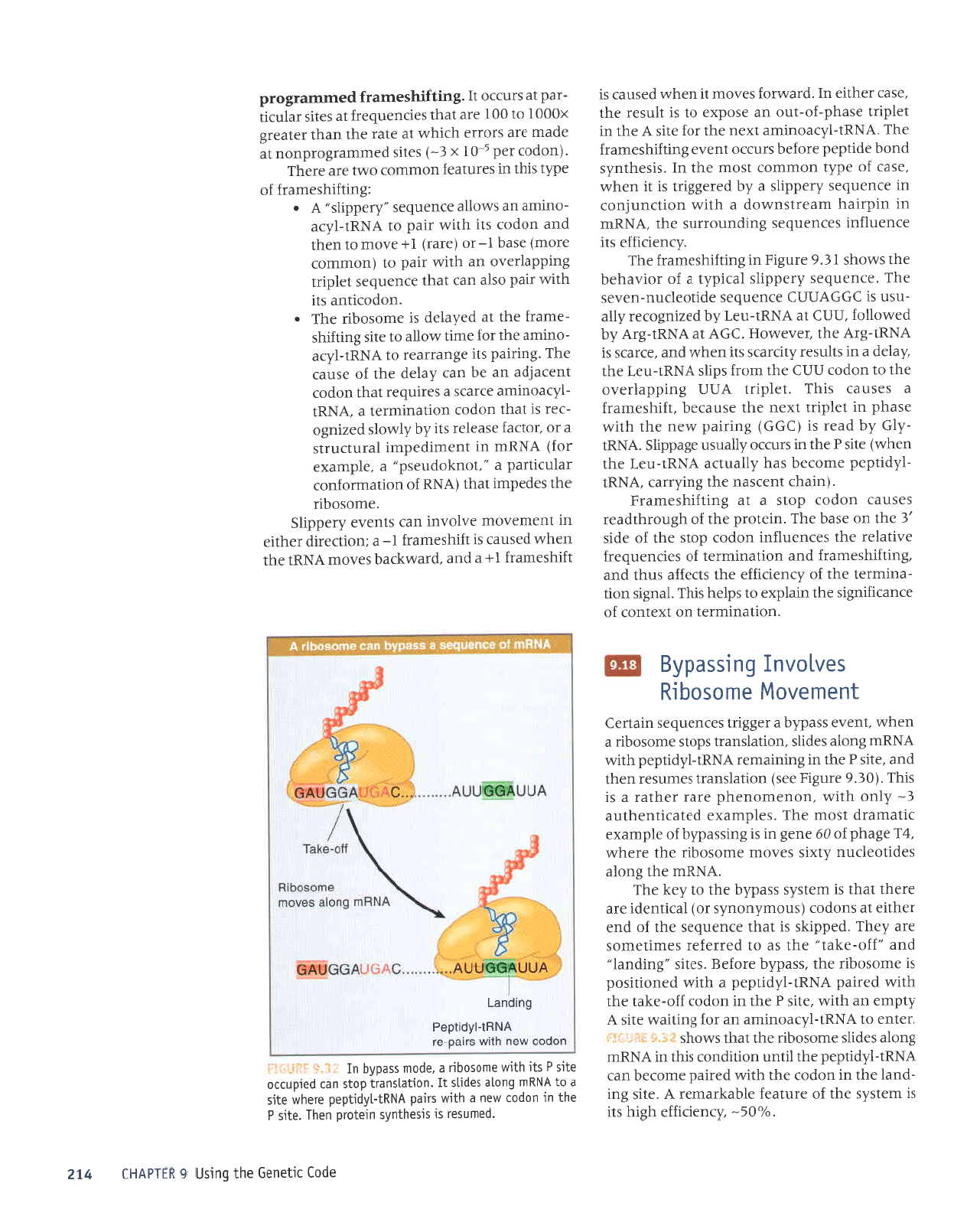
F5*tjftil *.h-* A IRNA that stips one base
in
pairing
with
a codon causes
a frameshift that can suppress termina-
tion.
The
efficiency
is usua[[y
-5%.
agGti** 9"3* Bypassing occurs when the ribosome moves
atong
mRNA
so that
the
peptidyt-tRNA
in the P
site
is
reteased from
pairing
with its
codon
and
then
repairs with
another codon
farther
alonq.
P
site.
The sequence between
the two
codons fails to be represented in
pro-
tein. As shown
in F3fitiRF.
+.3{1, this
allows translation to continue
past
any
termination codons in the intervenins
region.
60 nucleotide bypass
in
phage
T4
gene
60
Last codon
in
original
reading frame
odon in new
reading frame
Reading
without frameshift
GAUGGAUGAC............AUUGGAUU
Reading
after
frameshift
GAUGGAUGAC...,......,.AUUGG
212 CHAPTER
9

€rz
saluanbas
r{laddrts
le
slnllg
6ur4rqsauerl
11.6
pJIIe)
sr srqJ
'uortelsupJt
leJnteu
ur dats
.dressa
-JJu
p
se
uo
pJrTeJ
Jq
ueJ sluJle
,,3urpearsrtu,,
Jo
JJuJrrnf,ro (a1qellpa;d
1nq)
JreJ
Jr{t
tpql
lurod
tuelrodurr
aql
sJ>lelu
suorlenlrs qJnS
'auaB
q[1
tuanbasqns
Jqt
ppJr
ol lJpro
ur
t;rqseruer;
e.dq
passed,{q
Jq
tsnru
r,{7;o
uopor
uorteuruJal
Jql
qJIqM
ur
'tuJruJle
{J.
tsea,{
eqt
Jo
uortpnUs
Jelr
-rurs
Jr{l
sJlerlsnru
I
:
.,
,,
:
,
'SurSeruep
3q uel
.druaor;;a
ur eseJJJur
ue lsnlrl
aql
Jo
,{8o1orq
aqt
ur
tueuodurr
sr srqt'DpJ
uI'o/os-
z(1e;rd,{1
'.&rol
sr
4lqsJuerJ
aqt
;o
.,{ruarJrJJJ
JqJ
.(surat
-ordL1o4
ro;
epoJ
sJue)
Iplr^oJlag
,€.eZUo\)aS
osle
pue
67'6
an8qaas)
uopor
uortpurruJet
Jql
sasseddq
eJoJJJaqt pue
JueJJ
Supear puo)as
Jql
olur
sa8ueq)
tpql
UrqsJruerl
p
Lq
palelsuen
sr
(sasnrr,t
aruos
ur)
pup
'erueJJ
Supear
tuJJeJJrp
e
ur sarl
aua8
puof,Js
JriJ
'JrupJJ
Supear
Jqt
qlrM
aseqd
ur uopoJ
esuJsuou
e .{.q
paleurrural
sr
auaB
lsrrJ
Jql
Jo
uorlelsueJl
'sJSnrr^orlaJ
uI
'VNUur
eqt
Io
sJuJJdord
rrsurrtur
aql
ruoJJ
llnsal
pue
'srseqlu.,(s
uralord
Jo
uorteurur
-JJl
Jo
uollPnultuo)
Jqt
DJJJP
zieru
stuana qrng
'sJSn-rrA
pue
sa8eqd
Lq
paluasard
JJe
luJle
Ieru
-rou
e
sr Surl;rqsaruerJ
qJnIM
ur
suorlenlrS
'uopor
6ry
allpls
e
Aq paanol
-1o1
ete
(s)uopol
nol oml
qlrqm
lp
atuenbes
ospq-/
p
le
slnllo
lJtqs
oql
']uauala
r\1
lsear\
eq1
1o
aua6 q/Q
eql
Jo
uorssardxa
ro1
parrnbat
sr
Urqseup.U
l+
V
,,r
,i.
'uopoJ
P
purJ
ueJ
vNul
lxJu
eq1 JroJaq
paddqs
aq ol
espq
euo sJtroJ
srqJ
'pJ>lJolq
sr eseq
tuarefpe
Jql-JJupJpurq
rrrats
d1a4t
lsour-uospJJ
rJqlo
auos roJ
lnq
'uopol
JSeq-€
e sazruSolar
.{lqeqord
vNUt
Jos
-sarddns
JqJ
'salnJ
JIqqoM
Iensn
aql {q
patela.r
lou
JJe
,,uopoJ,.
ra8uol
eqr
Jo
uortrsod qunoJ
aql ur alqeldJJJp
eJe
teql
sJspq
Jlrleurelle
aql
'sespJ
aseql uI
'lup^JlalJr
sr JSpq
lxau
eql
1suor1
-rsod
aarql
tsrr;
er{l ur
f,)v
qtlM
,,uopoJ,,
JSeq-7
Lue
pear
uer
rossarddns
JJqlouV
'VvV
uopo)
Iensn
Jqt
Jo
ppJlsur
'nvvv
Jo
vvvy
JJqlrJ
ot
puodsar
uet ,rossarddns
s^lVNUt
IprJalJpq
e
'aldruexa
Jod
,,'uopoJ ,,
eseq-V
euo
uprll Jroru
azruSola:
uer
srossarddns
4rqsarueJJ
eruos
,,'uopoJ,,
JsPq
-7
e sazruSoJrr
VNUI
rossarddns
e{J
._+)f,))
aruanbas
taldnrpenb
Jql
01
+f,))
aluanbas
taldrrl
Iensn
Jq1 ruorJ
uopoJrlu€
aql
Surtral
-uor
'doo1
uopoJrlue
slr ur
pJuJsur
aseq
eJlxJ
ue
seq
leql
^reYNUl
e sr rossarddns
];rqsaruer;
Jr{J
'saspq
9
snon8rluoJ
IeJJAes
Jo
unr
p
ur
pelJesur
aq z(eur
g
e
'alduexa
Jod
.sJnprsJJ
Ie)rluJpr
Jo
r{JlaJls
e urqlrM
aspq
Ieuorl
-IppP
ue Sutuasur
dq
pasner
uJJq
seq
uortelnru
p
uJqM
atupJJ
Surpear
Jql slJJJJoJ.rossa:ddns
UlqserueJJ
leulJtxa
Jo
ad.{l
lsaldrurs
aq1
'saruadord
lupJrsqe
qll^\
svNul
Jo
turoJ
rql
ur
punoJ
eq ue)
osle
srossarddns
l;rqsaruer;
rrua8erlxa
JaAJ,lroH'(ta1dq1
sI epof,
Jlteueg
JqJ
,g.Z
uorl
-rag
aas)
aua8
e urqlrM
suoruasur
pue
suorl
-a1ap
aseq
Surtesuadruot
,{q palarqre
Jq
upJ srqJ
'JrrrerJ
Surpear
leufuo
aq1
Surrolsar,{q
passard
-dns
aq
uer z(aq;
'Jseq
e
Jo
uorlJlep
Jo
uorl
-Jasu
eql ruoJJ
llnsJJ
sluplnlu
ulqselupJd
.JIIS
Y
JqI UI
VNUIU
uMop ro
dn aseq
p
Jlotu
01
VNUI
e ,trolle
saluanbas
,,,{raddr1s,,
urplJJ)
.
.sJspq
aeJrll pnsn
Jql
Jo
ppalsu
saseq JnoJ
JoJ
,,uopol,,
e ezru
-3orar
srossarddns
VNUI
luelnrrr
JruoS
.
:seJuplslunJJrf,
o^1'l
Ur
s17Ngl
rqnads qll,lr
pJlprJosse
sr
Bur4lqsJurerd
'6ur1;rqsauetl
pau
uetbord
1o
eruaunrro
leln6at
eq1
uodn
spuadap
seua6
auos
Jo
uorlplsuetl
r
'aLuerl
6uLpeat
aq1 6u16ueqr
fqaraql
'uopolrlup
sl! ql$ parted
seq
lr
lalJp
oseq
auo fq
ryrqs
ol
VNUI
e
/1
ollp
saluanbes
&eddrt5
r
'luouruotnua
leuosoqu
aql
pup
VNUUI
Jo
eruanbes
eq1 fq
peruenllut
aq r\eLu
auet;
6urpear
aq1
o
saluanbo5
Areddq5
1e
srnllO
6uq;rqseuuJ
sr
rurs^s eq,
Io
alnlel; 3i;,fi"1i:l'#
:1,:1:i
-puel
eqr
ur uopoJ aqr
qllM parted
JruoJJq ue)
vNut-1^dppdad
aqt
[tun
uolllpuo) slqt ul
YNuru
Suop
sap11s
eruosoqrJ eql
lpql
smoqs
,
r:
:,,ri':,:;: ,
'JaluJ
ol
vNur-l.drBoulrue
ue ro; 8ur1te,u
alts
y
Ltdrua up
qtrM
'elrs
d
eqt
ur uopoJ
JJo-e>lpl
eql
qtyll
parred
VNut-lr{pudad
e
qlr,rzr pauorltsod
sr aurosoqrr
aqr
'ssed.{.q
eJoJJg'sa1ts
,,3utpue1,,
puP
,,Jlo-e>let,,
JVl se o1
perJaIJr
sJrullauros
are ,{aq1
'paddqs
sI
teqt
aruanbas aqt
}o
pue
JJqtre
te
suopof,
(snoru.{uouLs
ro)
pruuapt
are
Jraqt
teqt
sr ruats{s ssedLq
aql 01
,{a1
aq1
'vNuul
Jrll EuolP
sJprloel)nu
u(lxts sa.r.oru
JruosoqlJ eql
JJeqM
'71
a8eqdyo
99
aua?ur st Sutssedzi.q;o
alduexa
Jrlpruerp
tsoru
aqJ
'seldruexa
pJleJltuJqln€
g-
Lluo
qlIM
'uouaruouaqd
eJeJ JeqtpJ
e sI
slqJ
'(0€'6
arn8rg aas) uotlelsuerl
seunser
uJql
pue
'atrs
d
eql
q
Surureruar
yNut-1,{ppdad
qtu,r
VNUru
Suop
sapls
'uopelsuert
sdols aurosoqtr
e
uer{,lr
'lua^e
ssedLq
e ra33tr1 saruanbas
uleuaJ
luaua^ol/\l
euosoqru
sa^loAuI
6urssed^g
'uolleulluJal
uo
lxaluoJ
Jo
aruerr;ru8ts
aql urc1dxa
o1 sdlaq stql'pu8ts
uotl
-eururrJl
aqt;o
,buatJIJJJ
eql slJe}le
snql
pue
'3ur1;rqsarueJJ
pue
uollpulurrel
;o
satruanbarl
J^rleler
Jqt
sJJuJnlJuI
uopoJ dors
aql
Jo
JpIs
,€
eqt uo
Jseq JqI
'utalord
aqr;o
q8norqtpeeJ
sasneJ
uopo)
dols e
1e
Surlltqsaruerg
'(ureqr
tuJJSpu
aql
SurLrrer'VNUI
-ll.ptldad
auo]eq
seq
l,1en1re
VNUI-nef
rql
uaq,r,r)
Jtrs
d
eql
uI srnJ)o
Llensn a8edd{5
'ygg1
-r(tC
,(q
pear
sr
(fCC)
Sutrted
,lrru aql
qlIM
aseqd
ur
1a1dtr1
lxeu
Jqt
JSnPteq
'tJlqsaurelJ
e
sasne)
slql
'1a1drr1
y3n
SurddPlrelo
eqt ol
uopo)
nn)
aql
uo.ty sdqs
VNUI-nJT
rql
.{e1ap
e ur sllnsal
ltorers
sll ueqM
pup
'aJJP)s
sI
VNur-3rV
eql
'rr^rMoH')OV
re
yNul-8rv Lq
pe.&\o[o]
'nn)
lE
VNut-na'I
Aq
pazruSorar L1e
-nsn
sr
f,1;Dvnn)
a:uanbas Jplloapnu-ueles
aq1
'aruanbas
Lraddrls
lertdzll
e
Jo
rolleqeq
rql sMoqs
1g'6
arn8rg ut Sut4tqsaru€lJ
aql
'Aruatrt;;a
slt
aJuJnlJur
saruanbas
Surpunorrns
aql
'vNuru
ur urdrreq
rueeJlsumop
P
qll.l,r uorlrunfuor
ur aluanbas
Lraddrts e z(q
para88trl
sl
tI
ueqM
'esp)
Jo
ad.{.]
uoruuoJ
tsoru
aqt
q
'stsaqtu.ds
puoq
aprrdad
eroJaq sJn)f,o
tuene
3up;qsauerl
JqJ'VNUI-1.{leourrue txau
Jqt
JoJ elIS
V
Jq1 ul
1a1dut
aseqd-;o-tno
ue asodxa
01 sI
IFSaJ
aql
'esef,
JJqlIe
uI
'pJeMroJ
sa^olu
1I
uJqM
pJSnPJ
SI
apol
lr.laua!
aql 6urs1-1
'pounser
sr sLsaqlufs
uLelord
ueql
'a1ts
6
oql
ut uopo)
Meu
p
qltM
s.rred
y1511fpL1dad aleqm
alLs
p
ol
VNUtu
6uo1e
sapLls
1I
'uotlelsuP.ll
dols
uPl
patclnllo
alls
d
slt
q]tm
auosoqu
P
'apoul
ssedfiq
u1
r'
I
'' :'iii ril::
i
ulqseuerJ
I+
e
pue
'pJeM>peq
sa^olu
YNU1
Jql
uaqM
pasnPl sI
lJlqsaueJJ
I-
e
luoIDeJIp
raqlla
ur
luJurJnour
eAIoAuI
ue)
sluelJ
draddlS
'JruosoqII
aq1 sapadrur
teql
(VNU
Jo
uolleurroluoJ
relnrrl.red
e,,'tou>lopnasd,,
e'aldruexa
.roy)
ypgtu
ur
luarulpadul
lPJnlJnrls
e ro tolJPJ
esealer
s1r
Lq,{1a,ro1s
paztuSo
-Jar
sr
lPql
uopol
uolleulullJl
P
'VNUI
-1,{:eourue
J)lpJS
e sartnbal lPql
uopo)
luarelpe
ue aq
uPf,
,{.e1ap
aql
Jo
ssneJ
aq1
'3ur.rred
str
e8ue.trea;
ot
y1.trgt-1ri:e
-oulrup
eql
roJ erull
,lrolle
o1
alts Sutqtqs
-eue4
aqt
te
paLelep
sI euosoQlr
Jr{l
r
'uopoJIluP
sll
qtyrzr:red oslP
ueJ
]eq]
aruanbas
1a1drrl
3urdde1.ra,to
up
qlIM rted
ol
(uoruuror
aroru)
aseq
1-ro
(arer)
I+
eloru
01 uJql
pup
uopoJ
sll
qll,,ur .rred
ot
VNut-lAre
-oullue
up sMolle
aruanbas
,,Lraddqs,,
y
.
:bururqsaluerJ
Jo
adLl
sqt uI
setnleal
uourruol
oml
eJe
JJaqJ
'(uopor
rad
r-0I
x
g-)
salrs
paruurerSorduou
le
eperu
aJe
sJoJlJ
q)IqM
]e
aler
Jql
ueql
ralear8
x000I
ot
00I
aJe
teql
sJl)uJnbar; te
sJIIS
JPIn)p
-red
te
sJnlf,o
lI
'SuItJTqsaurEU
petutuerSo,rd
6
UltdvHl
,lz
uopoc
l ou
q11arr
srred-el
ypgl-1Ip;1de6
6utpuel

The
sequence
of
the mRNA
triggers
the
blpass. The
important
features
are the
two
GGA
codons
for take-off
and landing,
the
spacing
between
them,
a stem-loop
structure
that
includes
the
take-off
codon,
and the
stop
codon
adjacent
to the
take-off
codon.
The
protein
under synthesis
is
also involved.
The
take-off
stage requires
the
peptidyl-tRNA
to unpair from
its
codon. This
is followed
by a
movement
of the
nRNA
that
prevents
it
from
re-pairing.
Then
the ribosome
scans
the mRNA
until the
peptidyl-tRNA
can repair with
the codon
in the landing
reaction.
This
is followed
by the
resumption
of
protein
synthesis when
amino-
acyl-tRNA
enters the A
site in
the
usual way.
Like frameshifting,
the bypass
reaction
depends
on a
pause
by
the ribosome.
The
prob-
ability that
peptidyl-tRNA
will dissociare
from
its
codon in the
P site is increased
by
delays in
the entry
of aminoacyl-tRNA
into
the A site.
Starvation
for an
amino
acid can
trigger
bypass-
ing
in bacterial
genes
because
of the
delay that
occurs
when there is
no aminoacyl-tRNA
avail-
able to enter
the A
site. In
phage
T4
gene
60,
one role
of mRNA
structure
may
be to reduce
the
efficiency
of termination,
thus creating
the
delay that is needed
for
the take-off
reaction.
@
Summary
The
sequence of mRNA
read
in triplets
5'
-+
3'
is related
by the
genetic
code to
the amino acid
sequence of
protein
read
from
N- to C-terminus.
Of the
sixty-four
triplets,
sixty-one
code for
amino acids
and three
provide
termination
sig-
nals.
Synonym
codons that
represent
the same
amino acids
are related,
often
by a change
in
the third
base of the
codon.
This third-base
degeneracy,
coupled
with a
pattern
in
which
related
amino acids
tend to
be coded
by
related
codons, minimizes
the
effects of mutations.
The
genetic
code is universal
and
must have
been
established
very early
in evolution.
Changes in
nuclear
genomes
are rare,
but
some changes
have occurred
during mitochondrial
evolution.
Multiple
tRNAs
may respond
to
a
particu-
lar
codon. The
set of tRNAs
responding
to the
various
codons for
each amino
acid is
distinc-
tive for
each organism.
Codon-anticodon
recog-
nition involves
wobbling
at
the first
position
of
the anticodon
(third
position
of the codon),
which
allows some tRNAs
to recognize
multi-
ple
codons. AII tRNAs
have modified
bases,
introduced
by
enzymes that
recognize
targel
bases in the IRNA
structure.
Codon-anticodon
pairing
is influenced
by
modifications
of the
anticodon itself
and also by the context of adja-
cent
bases, especially
on the
3'side
of the anti-
codon.
Taking
advantage of codon-anticodon
wobble
allows vertebrate mitochondria to use
only twenty-two
tRNAs to
recognize
all codons,
compared
with the usual minimum of thirty-one
tRNAs; this is
assisted by the changes
in
the
mitochondrial
code.
Each
amino
acid
is recognized by a
partic-
ular aminoacyl-tnNl
synthetase, which also
recognizes
all of the tRNAs coding for that amino
acid. Aminoacyl-tRNA
synthetases
have
a
proof-
reading
function that scrutinizes the amino-
acyl-IRNA products
and
hydrolyzes incorrectly
joined
aminoacyl-tRNAs.
Aminoacyl-tRNA
synthetases vary widely,
but fall into
two
general
groups
according to the
structure
of the catalytic domain. Synthetases of
each
group
bind the IRNA from the side, making
contacts
principally
with the
extremities of the
acceptor
stem and the anticodon stem-loop; the
two types of
synthetases
bind tRNe from oppo-
site
sides. The relative importance attached to the
acceptor stem
and the anticodon
region for spe-
cific recognition
varies with
the individual IRNA.
Mutations
may allow a
IRNA to read dif-
ferent
codons; the most common
form
of such
mutations
occurs
in
the anticodon
itself. Alter-
ation of its specificity may allow a IRNA to sup-
press
a mutation in a
gene
coding
for
protein.
A
1RNA that recognizes a termination codon
provides
a nonsense suppressor; one
that
changes the
amino acid
responding to a codon
is
a
missense
suppressor. Suppressors of
UAG
and UGA
codons
are more efficient than those
of UAA codons, which
is
explained by
the fact
that UAA is the most commonly used
natural
termination
codon.
The efficiency of all sup-
pressors,
however, depends on
the
context of
the
individual target
codon.
Frameshifts
of the
+l
type
may be caused
by aberrant tRNAs that read
"codons"
of
four
bases. Frameshifts
of
either
+l
or
-l
may
be
caused
by slippery sequences
in nRNA that
allow a
peptidyl-tRNA
to slip
from its codon to
an overlapping
sequence
that can also
pair
with
its anticodon. This frameshifting also requires
another sequence that causes
the ribosome to
delay. Frameshifts determined
by the nRNA
sequence may be required
for
expression
of nat-
ural
genes.
Bypassing
occurs when a ribosome
stops translation and
moves along mRNA with
its
peptidyl-tRNA
in the P site until the
peptidyl-tRNA pairs
with an
appropriate codon;
then translation resumes.
9.19 Summary 275

References

Suppressors
May
Compete
with WiLd-type
Frameshifting
0ccurs at SLippery
Jakubowski,
H.
(1990).
Proofreading
in
vivo:
edit-
ing
of homocysteine
by
methionyl-rRNA
syn-
thetase in
E. coli. Proc
Natl. Acad.
Sci
USA 87,
4504-4508.
Nomanbhoy, T. I(.,
Hendrickson,
T. L.,
and
Schim-
mel, P.
(1999).
Tiansfer
RNA-dependent
translocation
of
misactivated
amino
acids to
prevent
errors
in
protein
synthesis.
Mol.
Cell 4,
519-528.
Nureki,
O. et al.
(1998).
Enzyme
structure with
two catalytic
sites for
double
sieve
selection
of
substrate.
Science 280,
578-58
l.
Silvian, L. F.,
Wang,
J., and
Sreitz, T.
A.
(1999).
Insights
into
editing from
an Ile-tRNA
syn-
thetase structure
with
tRNAIle
and mupirocin.
Science 285, lO7
4-1077.
Reading
of
the Code
Reviews
Atkins,
J. F.
(I991).
Towards
a
genetic
dissection
of
the basis
of triplet
decoding,
and its
natural
subversion:
programmed
reading
f rameshifts
and hops. Annu.
Rev.
Genet. 25, 2OI-228.
Beier, H.
and Grimm,
M.
(2001).
Misreading
of
termination
codons in
eukaryotes
by natural
nonsense
suppressor
tRNAs.
Nucleic Acids Res.
29,47674782.
Eggertsson,
G. and
Soll, D.
(
I 988
)
. Ttansfer
RNA-
mediated
suppression
of termination
codons
in
E. coli. Microbiol.
Rev.52,354-374.
Murgola,
E.
J.
(1985).
IRNA, suppression,
and the
cod,e. Annu. Rev.
Genet. 19,
57-80.
Normanly,
J. and Abelson,
J.
(
I 989).
Transfer
RNA
identity.
Annu Rev.
Biochem.
58, 1029-1049.
Resea
rch
Hirsh, D.
(197
ll . Tryptophan
rransfer RNA
as the
UGA suppressor.
J. Mol. Bi01.58,419-458.
Weiner,
A. M. and
Weber, K.
(1973).
A
single
UGA
codon functions
as a natural
termination
sig-
nal in the
coliphage
q
beta coat
protein
cistron.
J. Mol. Biol.80,
837-855.
The Ribosome
Inftuences
the Accuracv
of Translation
I(urland,
C. G.
(1992).
Translational
accuracy
and
the fitness
of bacteria. Annu.
Rev.
Genet.26.
29-50.
Ogle,
J.
M.,
and Ramakrishnan,
V.
(2005).
Sftuc-
tural
insights
into
translational
tidelity. Annu.
Rev.
Biochem. 7 4,
129-177.
Ramakrishnan,
V.
(2002).
Ribosome structure and
the mechanism
of translation. Cell
108.
557-572.
Resea rc h
Carter, A. P.,
Clemons, W.
M., Brodersen, D. E.,
Morgan-Warren, R.
J., Wimberly,
B. T.,
and
Ramakrishnan,
V.
(2000).
Functional
insights
from
the structure of
the
30S ribosomal
sub-
unit and its interactions
with antibiotics.
Nature
407,340-348.
Ogle, J. M., Brodersen, D. E., Clemons,
W.
M.,
Tarry
M. J.,
Carter,
A. P., and Ramakrishnan, V
(2001).
Recognition of cognate transfer RNA
by the 30S ribosomal
subunit. Science
292,
897-902.
Sequences
Reviews
Farabaugh,
P. J.
(1995).
Programmed translational
frameshifting . Microbiol. Rev. 60, lO3-134.
Farabaugh,
P.
J. and
Bjorkk,
G.
R.
(1999).
How
translational
accuracy
influences reading
frame maintenance. EMBO J L8, 1427-14]4.
Gesteland, R. F.
and
Atkins,
J.
F.
(1996).
Recoding:
dynamic
reprogramming of translation.
Annu
Rev.
Biochem. 65, 7 41-7 68.
Research
Jacks,
T.,
Power, M. D., Masiarz,F.R.,
Luciw, P. A.,
Barr,
P. J., and Varmus,
H. E.
(1988).
Charac-
terization of ribosomal
frameshifting in HIV- I
gag-pol
expression. Nature )31,
280-28).
Bypassing Invotves Ribosome Movement
Review
Herr,
A. J., Atkins,
J.
F., and Gesteland,
R. F.
(2000).
Coupling
of open reading
frames
by
translational
bypassing.
Annu. Rev. Biochem.
69,
)4)-)72.
Resea rc h
Gallant,
J.
A.
and
Lindsley, D.
(1998).
Ribosomes
can slide over and beyond
"hungry"
codons,
resuming
protein
chain
elongation many
nucleotides
downstream.
Proc. Natl. Acad. Sci.
usA95, t377t-t)776.
Huang,
W. M.,
Ao,
S.
2.,
Casjens,
S., Orlandi, R.,
Zeikus, R.,
Weiss,
R., Winge, D., and Fang, M.
(1988).
A
persistent
untranslated sequence
within bacteriophage T4 DNA topoisomerase
gene
60. Science
2]9, I005-1012.
Reviews
References 2t7
Protein Localization
CHAPTER OUTLINE
Introduction
Passage
Across a Membrane Requires a SpeciaI
Apparatus
.
Proteins
pass
across
membranes
through specialized
protein
structures embedded in
the
membrane.
.
Substrate
proteins
interact
directty with the transport
appa-
ratus of the ER, mitochondria.
or chtoroplasts,
but require
carrier
proteins
to interact with
peroxisomes.
e
A
much
larger and comptex apparatus is required for trans-
oort
into
the nucleus.
tfaEf
Protein Translocation May Be Posttranslational
or CotranslationaI
.
Proteins
that are
imported into
cytoplasmic organettes are
synthesized on
free ribosomes
in the cytosot.
o
Proteins
that are
impoded
into the ER-Gotgi system are
synthesized
on
ribosomes
that are associated
with
the
ER.
.
Proteins associate
with
membranes
by
means
of specific
amino acid sequences
calted signaI sequences.
.
Signal sequences
are
most
often leaders that are located
at the N-terminus.
o
N-terminaI
signaI sequences are usuatly cleaved off the
protein
during the insertion
process.
tEE
Chaperones May Be Required for Protein Fotding
.
Protejns
that can acquire their conformation spontaneously
are said to self-assembte.
o
Proteins
can often assemble into
atternative structures.
.
A chaperone
directs a
protein
into
one
particular
pathway
by excluding atternative
pathways.
.
Chaperones
prevent
the formation
of
inconect structures
by interacting with
unfolded
prote'ins
to
prevent
them from
fotding incorrectty.
Chaperones Are Needed
by NewLy Synthesized and by
Denatured Proteins
.
Chaperones act
on
newly
synthesized
proteins, proteins
that
are
passing
through membranes,
or
proteins
that
have
been
denatured.
o
Hsp70
and some associated
proteins
form
a
major
class
of chaperones
that act
on
many
target
proteins.
r
Group I and
group
II
chaperonins are large oligomeric
assemblies that
act on target
proteins
they sequester in
internaI
cavities.
.
Hsp90
is a
speciatized chaperone that acts on
proteins
of
signaI transductjon
pathways.
278
The Hsp70 Famil.y Is Ubiquitous
o
Members of the
Hsp70 fami[y are
found in
the cytosol,
in
the
ER,
and
in mitochondria and
chloroplasts.
.
Hsp70 is a chaperone
that functions on target
proteins
in conjunction
with DnaJ and GrpE.
SignaI Sequences
Initiate
Transtocation
r
Proteins
associate
with the
ER system on[y cotranstationatty.
.
The signa[ sequence
of the substrate
protein
is responsibte
for membrane association.
The
Signal
Sequence
Interacts with the SRP
.
The signaI sequence binds
to the SRP
(signaI
recognition
paticte).
.
Signa[-SRP binding
causes
protein
synthesis to
pause.
r
Protein synthesis
resumes when the SRP binds to the SRP
receDtor in the membrane.
.
The
signal sequence
is cteaved
from
the translocating
pro-
tein by the signal
peptidase
located on the
"insjde"
face
of the
membrane.
The SRP
Interacts with the SRP Receptor
r
The SRP is a comptex of
75 RNA with six
proteins.
.
The bacteriaI equivalent
to the SRP
is
a comptex of 4.55
RNA
with two oroteins.
o
The
SRP
receptor is a dimer.
r
GTP
hydrolysis releases the SRP
from
the SRP
receptor
after
their interaction.
The
Translocon Forms a Pore
e
The Sec61 trimeric
complex
provides
the channel
for
proteins
to
pass
through a
membrane.
o
A transtocating
protein passes
directly
from
the
ribosome
to the translocon
without exposure to the cytosot.
Transtocation Requires
Insertion into the Translocon and
(Sometimes)
a Ratchet in the
ER
o
The ribosome,
SRP, and
SRP receptor are sufficient to insert
a
nascent
protein
into a transtocon.
.
Proteins
that
are inserted
posttranstationatty
require addi-
tionaI components
in the cytosol and Bip in the ER.
.
Bip is a ratchet that
prevents
a
protein
from
stipping
backward.
Reverse Transtocation
Sends
Proteins
to the Cytosol
for Degradation
.
Sec61 transtocons
can be used for
reverse
translocation
of
proteins
from
the ER into
the cytoso[.
Proteins
Reside in
Membranes
bv Means
of
Hydrophobic
Regions
r
Group I
proteins
have
the N-terminus
on
the
far
sjde
of the membrane; group
II
proteins
have
the opposite
orientation.
.
Some
proteins
have
multipte
membrane-
spanning
domains.
Anchor
Sequences
Determine
Protein
0rientation
o
An anchor
sequence
halts
the
passage
of
a
protein
through
the
transtocon.
Typi-
catty this is
located
at the C-terminaI
end
and resutts
in
a
group
I
orientation in
which
the
N-terminus
has
passed
through
the membrane.
r
A combined
signat-anchor
sequence
can
be
used to insert
a
protein
into
the
mem-
brane and anchor
the
site of insertion.
Typicatty
this
is
internaI
and results
in a
group
II orientation
in which
the
N-
terminus is
cytoso[ic.
How Do Proteins
Insert
into
Membranes?
r
Transfer
of transmembrane
domains from
the transtocon
into
the tipid
bitayer is
triggered
by the interactjon
of the
trans-
membrane
region
with
the translocon.
PosttranslationaI
Membrane
Insertion
Depends
on Leader
Sequences
o
N-termina[
leader
sequences
provide
the
information
that altows
proteins
to associ-
ate with mitochondriaI
or
chtoroolast
mem
branes.
A
Hierarchy
of Sequences
Determines
Location within
0rganelles
e
The N-terminal
part
of a leader
sequence
targets
a
protein
to the mitochondrial
matrix
or chloroptast
[umen.
o
An adjacent
sequence
can control further
targeting
to a
membrane
or the intermem-
0rane
sDaces.
r
The
sequences
are cleaved
successivety
from
the
protein.
Inner and
0uter Mitochondrial
Membranes
Have Different
Translocons
o
Transpoft
through
the
outer and inner
mitochondriaI
membranes
uses different
receptor
comptexes.
e
The TOM
(outer
membrane)
complex
is
a
large
comptex
in which
substrate
proteins
are directed to the
Tom40
channel by one
of two subcomptexes.
r
Different TIM
(inner
membrane)
complexes
are used depending on whether the
sub-
strate
protein
is targeted to the
jnner
membrane or to the lumen.
r
Proteins
pass
direct[y
from
the
TOM
to the
TIM comp[ex.
Peroxisomes
Employ Another Type of
Translocation
System
o
Proteins are imported into
peroxisomes
jn
their futly fotded state.
o
They have
either a
PTS1
sequence at the
C-terminus or a
PTS2
seouence at the
N-termi nus.
e
The receptor Pex5p binds the PTS1
sequence and the receptor PexTp binds
the
PTSZ
seouence.
o
The receptors
are cytosolic
proteins
that
shuttle
into
the
peroxisome
carrying a
substrate
protein
and then
return
to the
cytosoL.
Bacteria Use Both CotranstationaI
and PosttranslationaI Translocation
.
Bacterial
proteins
that
are
expoded to or
through
membranes
use both
posttransta-
tionaI and cotranslationaI
mechanisms.
The
Sec System
Transports Proteins into
and Through the Inner Membrane
.
The
bacteriat SecYEG
translocon in the
inner membrane is retated to the eukary-
otic
Sec61
translocon.
r
Various chaoerones are
invotved in
direct-
ing
secreted
proteins
to the trans[ocon.
Sec-Independent
Tra nstocation
Systems
in
E. coLi
t
E.
coli and organetles
have retated systems
for
protein
transtocation.
.
One system attows certain
proteins
to
insert into membranes without a trans[o-
cation apparatus.
r
YidC is homologous
to
a mitochondrial
system for transferring
proteins
into
the
inner membrane.
r
The
tat system transfers
proteins
with
a
twin arginine
motif into the
periplasmic
sDace.
Summarv
CHAPTER 10
Protein Localization 219
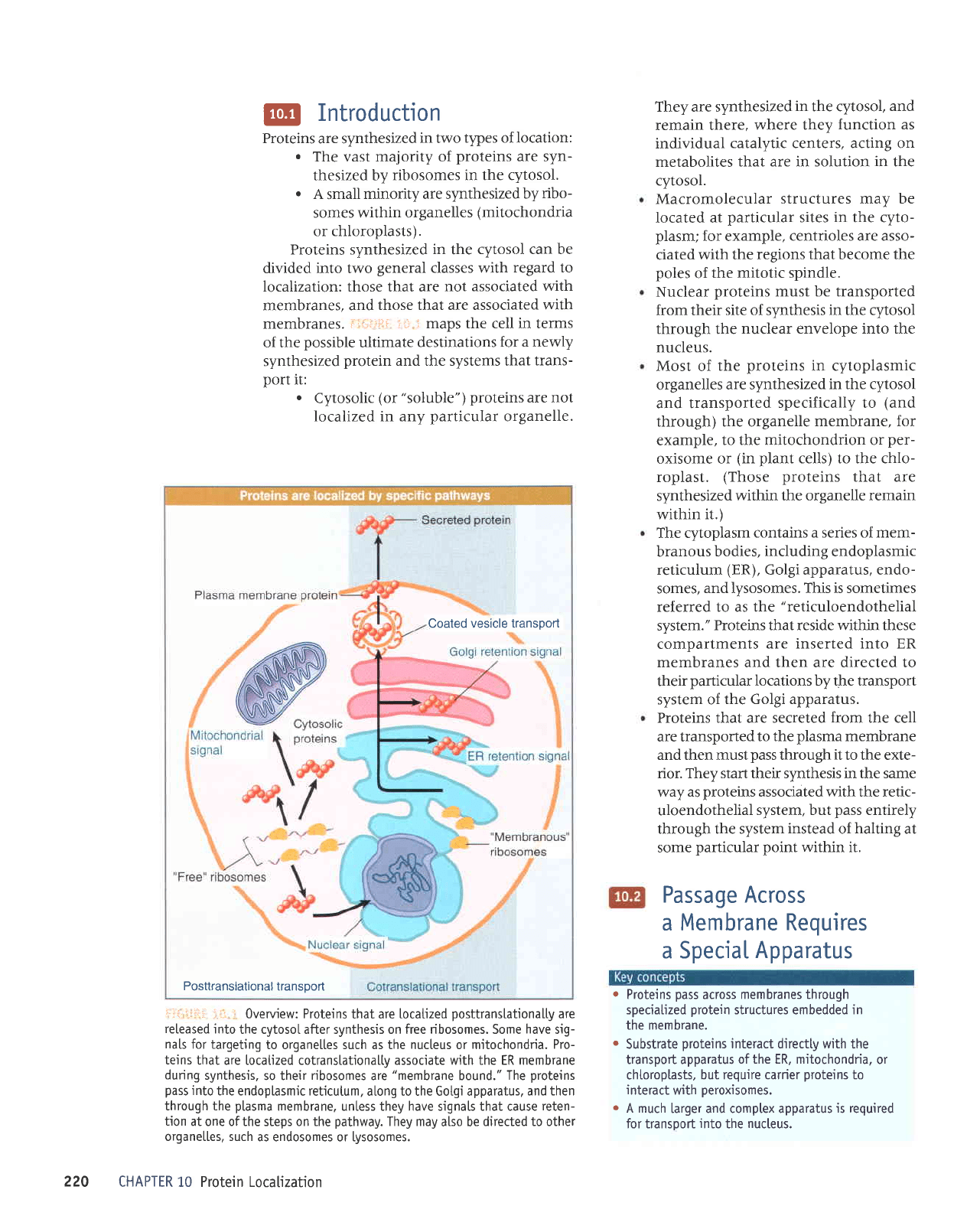
@
Introduction
Proteins
are synthesized
in
two
tlpes of location:
.
The vast majority of
proteins
are syn-
thesized by
ribosomes in the cytosol.
.
A
small
minority are sl,nthesized
by
ribo-
somes within organelles
(mitochondria
or chloroplasts).
Proteins synthesized
in
the
cytosol can be
divided into two
general
classes with
regard to
localization: those that are
not associated
with
membranes, and
those that are associated
with
membranes.
i-i:;ir::i ::J:
:
maps the cell in terms
of the
possible
ultimate
destinations
for a newly
synthesized
protein
and the systems that
trans-
port
it:
.
Cytosolic
(or
"soluble")
proteins
are not
localized in any
particular
organelle.
J;.::
r.::::
i i-t
:
Overview: Proteins
that are
locatized
posttranslationalty
are
released
into
the cytosoI after synthesis on free ribosomes. Some
have
sig-
nals
for targeting
to organeltes such as the nucteus or mjtochondria.
Pro-
teins
that are locatized cotranslatjonalty associate with the ER membrane
during
synthesis,
so their
ribosomes
are
"membrane
bound."
The
proteins
pass
into
the endoplasmic reticutum,
a[ong to the Gotgi apparatus,
and then
through the
plasma
membrane. untess they have signats that cause
reten-
tion
at one ofthe steps on the
pathway.
They may
also be
directed to other
organe[[es, such
as endosomes or lysosomes.
CHAPTER 10 Protein Localization
They are synthesized
in
the cytosol,
and
remain there,
where they
function as
individual
catalytic centers, acting on
metabolites
that are in solution
in
the
cytosol.
Macromolecular
structures may be
Iocated at
particular
sites
in
the cyto-
plasm;
for example,
centrioles are asso-
ciated
with the
regions that become the
poles
of
the mitotic spindle.
Nuclear
proteins
must be transported
from their
site of synthesis in the cytosol
through the
nuclear envelope into the
nucleus.
Most of
the
proteins
in cytoplasmic
organelles
are synthesized
in the cytosol
and transported
specifically to
(and
through) the organelle
membrane, for
example,
to the mitochondrion or
per-
oxisome or
(in plant
cells) to the chlo-
roplast.
(Those
proteins
that are
synthesized
within the organelle remain
within
it.)
The cytoplasm
contains a series of mem-
branous
bodies,
including
endoplasmic
reticulum
(ER),
Golgi apparatus, endo-
somes,
andlysosomes. This is sometimes
referred to as the
"reticuloendothelial
system."
Proteins that reside within these
compartments
are inserted into ER
membranes and then are directed to
their
particular
locations by the transport
system of
the Golgi apparatus.
Proteins that are secreted from the cell
are transported
to the
plasma
membrane
and then must
pass
through
it
to the exte-
rior. They start their s)'nthesis in the same
way
as
proteins
associated with the retic-
uloendothelial
system, but
pass
entirely
through the system
instead
of
halting
at
some
particular
point
within
it.
a
a
Coated vesicle transDort
Posttranslational
transDod
Passage Across
Membrane Requires
SpeciaI
Apparatus
r
Proteins
pass
across
membranes through
specialized
protein
structures embedded in
the membrane.
.
Substrate
proteins
interact directty with
the
transport apparatus
of the ER, mitochondria,
or
chtoroptasts,
but require carrier
proteins
to
interact
with
peroxisomes.
r
A much
larger and comptex apparatus
is required
for transport
into
the
nucteus.
220
