Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.

tLl
auosoqru
aq] o] Alalpurallv
pulg
slollpj
uoqebuoll
EI.g
'alelolsuerl
puP
'spuoq
0pt1
-ded
urol'ylilXllAreouLLue
mau
ldacre
sauosoqu se
saleu
-lallP
9-ll
puP
nl-ll
slolleJ
Jo
6urpurg
i
. i:t
,tlitlii:1
'lleuruaq
'enqJpV
Jo
^lrsle^run
'uessrN
1no3;o
fsalnor
oloqd
'aldrnd
ur ale
g-ll
uL
1L
6uLlquLaser
uteur0p
oql
pup
VNUI
aq1 luearO
pue
pol
ur
atp
g-ll
pup
n1-ll
Jo
sureuop po^.rasuot
f11etnpnr15
'(1qbu)
9-13
1o
alnJlnrls
oql salquasol (Uat
) a.ll-nf
-11-y11
gllfreourue
1o
xalduior
[reutal
aql
Jo
alnl]nl]s
aLlf
i.rr
::
iirt;
;i:]
Suraq;o
ppelsut
JruosoqrJ
aql uo urprual
d(C
pue
5)-dg
lPql
os
'qSnoqr'xaldruor
doD-t-da
-aruosoqrJ
Jql sezllq4s
pr]p
Jrprsnd
'sapuoelJnu
eJJql sJ^oru
JrrrosoqrJ eql
pup
'paz,{lorpdq
sl
dI9
'Jruosoqrr
Jql 01 spulq
D-d!I
:srnJJo
uorleJolsuell
Jo
punoJ
Juo
'prJe
trprsnJ
Jo
J)ue
-sard
aql uI
'(;.
t
'i:
:J;lii
ilii
I
ees) elP1s uorlpJolsuerl
-lsod
st1 ur JruosoqrJ er{t
,,srue[,,
qJrqM
'pr]p
rrprsnl
Jltorqrlue
prorets
aqr
Jo
slJJJJr
aqt
^q
paJa^oJsrp
se1!t JSealaJ
D-dg
JoJ
pJJu
aqJ
'D_dg
JSPelar
o1
prprru
sl
dJC
Jo
srs^loryIq
rqJ'(srs^lorpdq
afC
JO
sJuJSqe
JrIl Ur JaMOIS
qJnru
Sr UOrle)OISUeJI
q8noqtle)
uortef,olsuprl
JoJ
lertuessa
.{latnlosqe
lou
sr srs,,[.1orp,{q
s1r
tnq
'Surpurq
JoJ
pJpJeu
sI aplloJlrnu euruen8
p
Jo
Jf,uesJJd aqt snql
jdJ)
roJ
prtrupsqns
sl
d)d-dw)
uJqM euosoqrr
Jql ol
pulq
IIIIS
ueJ
f,)-dEI
'luJruJ^our
eruosoqrJ
Surrttollo;
pJseJIJJ
sr uJql
pue
'uorlelolsueJl
rosuods
ol eruosoqrJ Jql ot spulq
C-{f
'uorlf,unJ
str
IIIJInJ
ot spreu
rl tpql
dJg
aql
;o
.duedruot
Jql uI,{po aurosoqrJ
Jql 01 sseJJP surplqo
Jol
-rPJ
qJPe
leql
seJnsuJ
qJIqM
'JrxosoqrJ
Jql ol
Surpurq ro;
parmbar
sr uJoJ aleqdsoqdr.rl
aq1
ac9
01
punoq
ueqM J^rlfeur
lnq
al{)
o1
punoq
uJqM alrlJe JJp
teqt
suralord Surpurq-41g
)rJJruouoru JJe
sJolJeJ uorle8uola
qlog
'reuuPur
AIJepro
ue ur
peaJord
srsaqluLs uralord
Jo
sluela
Jql
leql
seJnsuJ
purq
ueJ rJqlo
aql aloJaq
psseJIJI
eq ol ropeJ
qJea
roJ
paau
eqJ
'(etls
v
aqr
Jo
^1r
-urJrl
Jql ur,{.lqerunsard)
alrs Surpurq erups
aql
roy aladuor ^Jqr
leql
uorldrunsse JterpJrurur
Jql saleJJ) slqJ
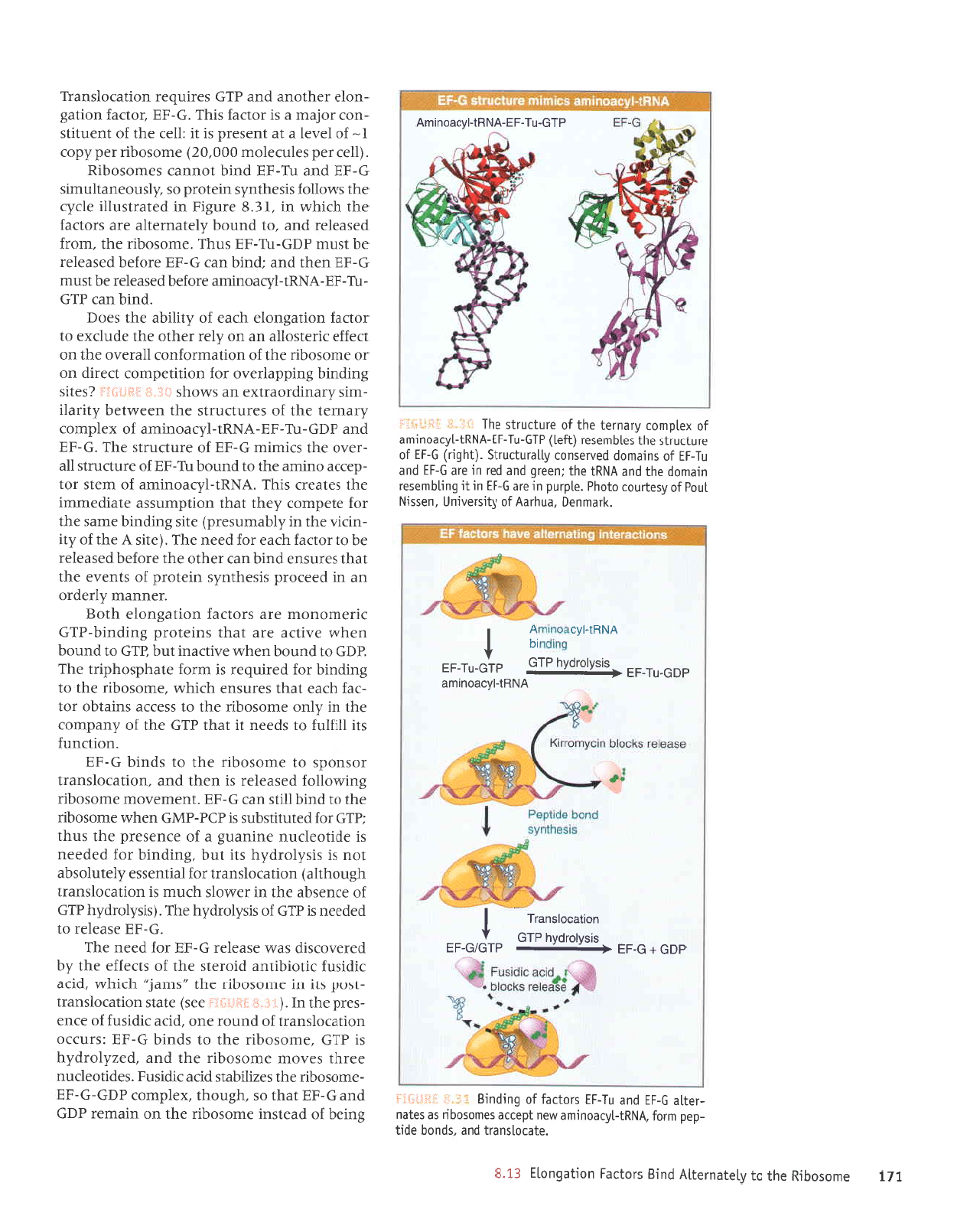
'vNul-12(reou[ue
]o
tuJls Jol
-darre
ourue
Jr{l 01
punoq
nJ-llg
Jo
JJnlJnrls
[e
-JeAo
eril sJrrurru
c-!Is
Jo
eJntJnJts
aqI
'c-!Ia
pue
d(D-nJ-{iI-VNUI-l^f,eourrue
yo
xalduor
.{*teu.tal
Jql
Jo
sJJnlJnJls Jqt uJJMtJq ,(1r:e1
-ruls
^JeulpJoerlxe up smoqs
ii{'ij
iiii:l,i,liri
ZsJlrs
Surpurq
Surddelraao roJ uorlrtaduoJ
lJJJrp
uo
Jo eruosoqrJ eql
Jo
uo4euJoJuoJ
IIerJ^o
eql uo
tJJJJe
Jrrelsoflp
ue uo,{1ar raqto Jq} JpnlJXJ
o}
rotleJ
uolteSuola
qrea yo
,{111tqe
aql sJo(
'purq
uer
dJ9
-
nJ-{A-VNp-pbeourue JroJJq
pJspJIJr
aq
tsnru
g-49
uael
pue
jpurq
uer
{)-dA
eroJaq
peseelJr
eq
lsnu
d(Ic-nJ-d!I
snqJ
'Jruosoqrr
Jql
'urolJ
pJseelJJ pup
'ot
punoq.dlateuratp
aJp sJoDeJ
rql
qlrqM
ul
'I€'8
arn8rg
q paleJtsnlg
apLr
Jql suolloJ
srsaqtuz(s uralo;d os
,(lsnoauetlnurs
f,)-{!I
pue
nJ-!I[
pulq
louuEJ
saruosoqru
'(1ar
rad sJInJJIoru
000'02)
Juosoqrr rad ddor
I-
Jo
IJ^el
e
te tuasard
sr
tr
:ilel
Jqt
Jo
luJnlus
-uoJ
Joleru e sr ropeJ sIqJ
'D-dA
JopeJ
UoUBB
-uolJ
Jaqloue
pue
dIC
salnbar uorteJolsue.\,1
doe
+g-13
ffi
dle/g-ll
uorlpcolsupjl
I
vNUl-lAcPourure
doo-n1-r3
=-"6,p-,ffij5
.rf
-nr_r=
!ql
VNU}IAC
V
I
dl0-nI-lf
-vNHFl^c€oururv

srsaqlu^s
uralord
s
ultdvHl
zLl
('paralunorua
sr uopoJ
uorlPurrural JaqlouP
Irlun
srs3q1
-uls
uratord
Jo
uorlpnurluo) eql ul stlnsaJ
'1I
o1 spuodsar
.dpadordurr
yNdl-lzbeoullup
ue
uJr{,lr
'uopoJ
uorlpurrurel
e Surpear uI rorrJ
uV)
'vgn
Surpear sJoJre arou aq
o1 rBaddB
araql
q8noqlle
'Cvn
ueql
^lr^peq alolrr
pesn
sr
yDn
'uopoJ
uorlpunrrJJt
pJsn
Aluour
-uroJ
tsour
aq1 sr
vyn
'saua8
IelJelleq
uI
_
'a)uJnDes
Surpor
e urqlrm uoltplnur
^,{.q
patean
ro aua8
p
Jo
pua
eq1
le
,{11ernleu 3ur;:nlo raqtaqm
'srsaqluz(s
uralord
pua
ot
tualJrJJns
pup
z{.ressarau
aJoJJreql are saruanbas
laldtrr
y9n
pup
'VVn 'Dvn
aqJ'aua8 e urqlru srsaqlu,{s
uralord alpurruJal
ol
luJrJrJJns
sr suopoJ eaJql
aqt
Jo
euo
t(ue
teqt
Moqs
suoltptnlu
JsuasuoN
'af,uanbes
ad,{.r-ppa aql
Jo
prJe
ourure
Ipulu
-Jal-)
aql Sultuasardar uopo; aqt Jat;e
^lalp
-rperuurr
serl
suopoJ uorleurruJal eql
Jo
auo
'paruanbas
ueJq spq
teql
auaS
l:arra u1
('uopoJ
dols sr
ruJal JJllJq
y
'aua8
luelnru
e ur auo
arilldnrsrp e
f1aq1e-Suruparu
J^pq op
suopoJ
eql
lpql
ualt8
'raruouslru
e z(11eat sI
,,asuJs
-uoN,,
'sla1drr1
uorleurruJet Jqt aqrJJSJp 01
pesn
sr
uopu asuasu7rl
urJal eq]
sJrurlauros)
'uollelntu
asuasuotr P
pJIIPJ
sr
tros
srql;o a8ueqr
y
'(pa1ero1
sr alrs
luelnur
aqt uratord aqr Euole
reJ ,lroq
uo
'JSJnoJ;o
'Surpuadap)
uorpun; ural
-ord
qsqoqe
o1 ,(1a>1t sl sIqJ'llrr
tuetnru
aql ul Jperu
sI uleloJd aqt
;o
ued
rsrrl
aqr.{pg
'uopor
tuelnru
aql
le
slsaqtu.{s
uralord
Jo
uolleullural arnlBruard
sasneJ
1l
'suopoJ
uorlPurrrrJal aeJql Jq]
Jo
auo selpJJJ uorlelnur
lurod
e ueefi{ o
'luaurareldar
prJE
ourue Jq1
Jo
arnleu aql
pue
uorlelnlu
Jo
Jlrs aql uo
spuadap uorlJunJ uralord uo
DaJJa
eq1
luralord Jqt ut rJqlo
Jqt sareldar
pne
ourruP auo
'uorlPlnru
asuassF.u e
pa[P)
sr
prJe
ourure
luJJJJIrp
e
tuasard:r
o1
uopof,
e sa8ueqr
leqt
uorlelnru
lurod
y
.
:uorlelnru
rugod
yo
saddl
om1
paqsrn8uusrp
lpql lsal
rrlaua8 e ,{q umoqs
Lleut3rro se,u s1a1du1 Jseql
Jo
JJnlpu aqJ
'uopor
pdo
aqt
pallpJ
serupeuos sr
Vgn
puP
'uopoJ
alqJo Jql sr
vvn
'uopoJ
Jaqurc
Jq1
pJIIeJ
s1
la1drr1
Cvn
JqJ
',{.ranorsrp
rreqt
Jo
,{.ro1srq eql ruoJ} sJrueu
IenseJ
aneq Laql
'srs
-aqluz(s
uralord
pue
r{Jrqm
'(suopoc
dols
ro)
suopof, uorlpurrura]
are staldut eJJq] Jaqro eqJ
'sprJp
ourrue o1
pau8rsse
are s1aldrrl
I9
^luo
'9Vn<V9n<VVn
sauuanDalJ
e^rlplar
qlM^
ua1o
lsour
pesn
ate
feql Puapeq
uI
'srseqlufis
uralord
aleutural
V9n
pup'Qaqure)
9y1
'(aqro)
VVn
suopor aql
srsaqlu^s
uralord
alPururel
suopol
aalql
'IIJJ
P
III>I
01 seln)elou
zJge
luel)lJ;ns
z(;tpour
uP) ulxol
Jo
alnJJloru
a13uts
v
:alll)alla ,{1auar1xa
st
uorlJper
eqJ,'ulxol erraqlqdrp
Jo
slJJIJa
IPqlal
aql JoI alqtsuodsar
s1 uor1e1,{soqlr-d(V
eqJ
'sartads
,{.uetu
Jo
Zdga
aql
01 uoluluoJ
sI
ll
iaulppslq e Eud;
-poru
z(q
parnpord sI
lPql
pIJe
ouruP
lPnsnun
up sr
luauq)etlP
Jql JOJ
elPJlsqns
eqJ
'slseql
-uds
uralord ur anrDeul
st ateEnluor
Zdila-ud11v
eqJ
'Z{aa
rql oluo
l.latoru
(uaAV)
ldsoqtr
aleqdsoqdp Jursouepp
ue
JaJSupJ] 01
JoIJPJoJ
e se
(qy51)
aplloalJnulp
auluape
aplrueull
-oJru
sesn urxol
JqJ
'ulxol
elraqlqdtp
o1
,{1t
-UqlldeJsns
sll sI
Zd!IJ
Jo
uollf,Par
anbrun
y
aIO
sU;o
srsz(lorplq
luanbasuor
qltm
saruosoqrJ
01
pulq
uer
xalduo)
aq1
pup
'parPl
-osr
eq ue)
dJD
qllm
Zdga;o
xaldruor
elqels
V
'prJp
JrprsnJ ^q
pellqlqq
sI os1e
uoll)E s11
'sts,{1
-orpdq
dJg
uo
luapuadap
espf,olsuerl
e sP Jeu
-uetu
Jepturs P ur suorlJunJ
q)lqm'719a
uratord
Jql
sr
D-Ja
ol
updJalunor
ttto{re>1na
aq1
'llunqns
s05
3q1
prPl11tol palualJo
ppalsu
sr
t
urPuop
'uope)olsuPJl
JauY
'YNUI
aqt
Suneldslp roJ alqlsuodsar
aq
ppol
uletuop
srqJ'JIIS
V
aql orul
pJlJJsul
sI
qJIqM
'7
ulPurop
prllp)
uor8ar e
z(q aperu alp
tlunqns
S0€
aql
qlIM
sDeluoJ sll
Jo
lsow
'sllunqns
leluosoqlJ
o1l^1 aql ssoJJP
punoq
sI
1r'uoltPJolsuPrl
eJoJag
'uope)olsueJl
le
sJn)Jo
D-IIEI
io
uoIlPluaIJoJJ
a^rsuelxe uv'aJnlf,nrls
Jllrosoqu
aql ut aSueqr
e sa)JoJ
uJnt ur
qJIqM
'C-ca
Jo
JJnlJnrls
Jq1
ur a8ueqr
p
sasnp)
srszllorplq
dJD
lpql
sI Iuslu
-eqJeru,{1a111
tsoru
JqJ'lualualoru
JIIrosoqIJ
aql
sJleJaleJJp
puP
uolleJolsuPJl
eJoJeq
sJnf,Jo
qrlqm
'srsilorpz(q
41)
qtlm
uollJunfuor
uI
D-1IE
Lq
pale.Lrlrp
s,lr
'ralJ,lrog
'(sralua)
J^lllv
IpJeAaS
eApH saruosoqry
'Z
I'8
uoIlJeS aas) arnl
-JnJls
ur a8ueqr
rofeur e sartnbar
lpql
auosoqlJ
aqt;o
[lradord
rtsurrlut
up sI uolteJolsue4l
'ureqf,
eql o1
pappe
aq UPJ
spr)e oururp JequnJ
ou
pup
'VNg]-lzbeouture
pulq
louueJ
aruosoqlJ
aql
'lFsal
P sv
'psseeleJ
ELI
srollpJ
ur.olord [q pazruborau
erv suopo]
uotlputural
9I.g
(99'g
arnbtl
aas)
rolre; 6utt:Aral
oulosoqu
aql sl
JUU
'a1s
lpulosoqu
aurps
aql ol
pulq
0l
EJU-Z/IJU
slotleJ aspalal oql
pup
'!-ll
lope1
uorlplolsupll
aq1
'xalduor
VNUI-nl
lollpJ
uotle6uola
aql salqpua AllLturru
lelnrelo1,1
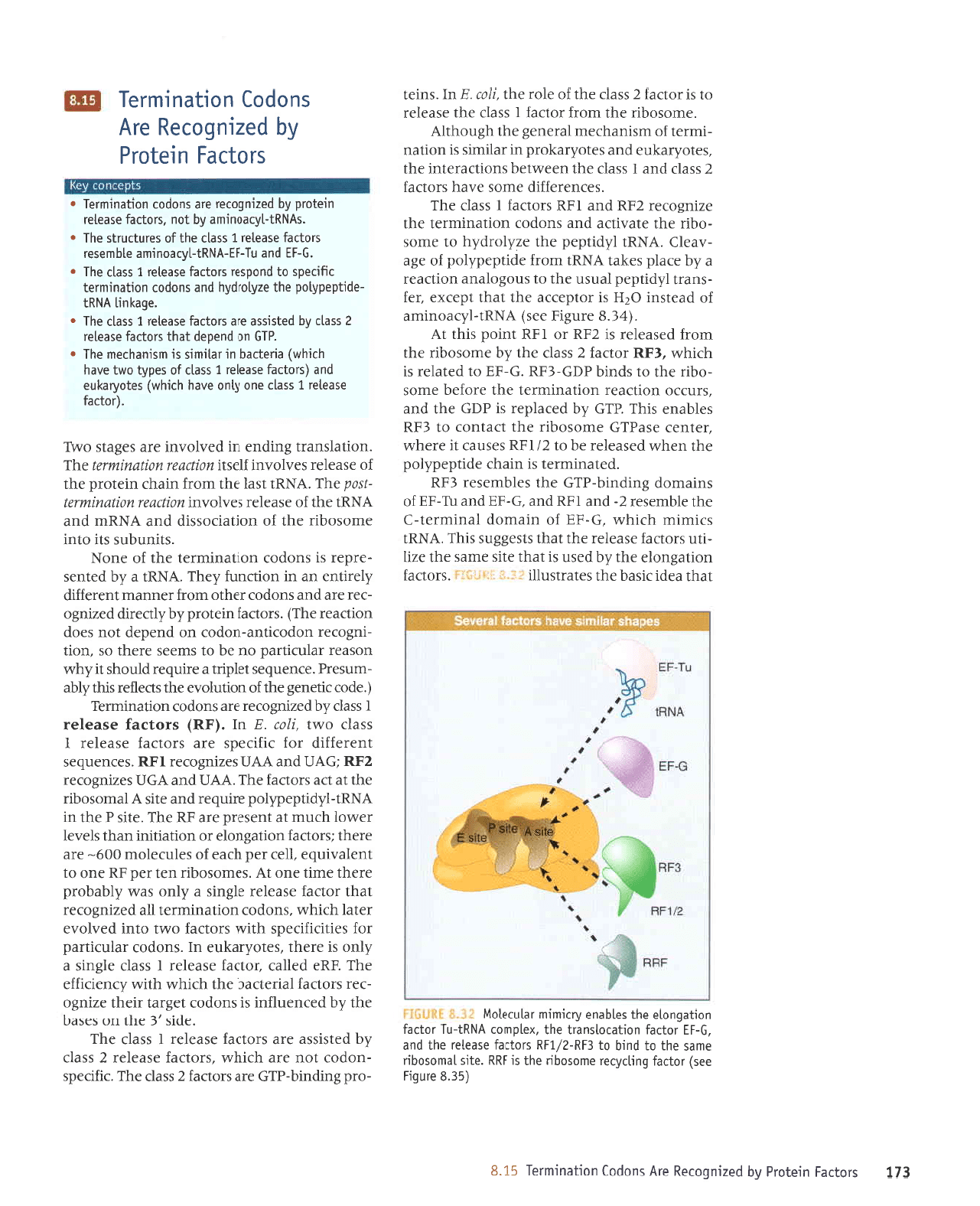
lEqleJpIJISeqeqlsJlPJlSnllI.
I
i,
ri,1,:'SJOIJPJ
uorleSuola Jql ^q
pasn
sr
]eql
aus aues
Jql ezrl
-rln
srotrpJ
aspaler rqt
teql
s1se33ns slql
'VNUI
sJrrurur qJIqM
'g-df
Jo
ureuop
IeurruJal-)
eql alquasrr
z-
pue
Idu
pup
'g-{f
pup
nJ-dll
Jo
sureuop
Surpurq-419
aqt selquJser
€JU
'peleurrurJt
sr
ureqf, aprldadz(1od
eql
uaqM
pasPJlar
aq 01
z/I{u
sesne)
1l
JJaqM
'ratue)
JsedJg
auosoqrJ Jqt
tJeluol
01
€Ju
salqeua slqJ
aJC
,
q
pareldar
sl
d61C
eql
pup
'srnJJo
uorlJeeJ uorleururJel
aql JJoJeq Jruos
-oqrr
rql o1
spulq
d(D-€du
')-dg
01
pelPlar
sr
q)lq,rr'€dU
rotlpl
Z
sselJ eql ^q JuosoqrJ aql
Iuorl
pJSeJIaJ
sl
ZIU
Jo
I{U
rurod
srqr
ry
'
(7E'g
arnSrg
aas)
yggt-1,(Jpourrue
Jo
peetsur
OzH
sI roldJJJe
aq1
leqt lda)xe
TJJ
-suer1
1^dprldad
lensn
aqt ot snoSolpup uorpeel
e ,{q areld
sa>lpl
yNUl
ruorJ
aprtdaddlod;o
a8e
-^prl)
'vNUt
IipltdaO
eql az.{1orp,{q 01 rruos
-oqu
Jql
eleAIlJP
pup
suopor
uorlpurrurJl
eql
azruSorar
ZdU
pue
I{U
srolJpJ
I
ss€lJ
eql
'saJuaJaJlrp
eluos e^er{
sJolJeJ
z
ssPIJ
pue
I
sselJ
eql ueearlJq suorDeJelq aql
'salo^J€>lnJ
pue
salo^dre>loJd
ur Jelrurs sr
uorleu
-rurrel
Jo
rusrupqJJur
leraua8
aqr
q8noqrly
'auosoqrJ
er{l ruorJ
Jope}
I
ssep eql
JSPJIaI
o1
sl JotJPI
z
sselJ eql
Jo
alor ertrl'!ln
'!'
q
'sulal
-ord
Surpurq-dJg
Jre sroDeJ
Z
ssplJ aq1
'rryoads
-uopol
lou
JJe
q)lqM
'sJoDPJ
aSPJIJJ
z
sselJ
,{.q
patsrsse
Jrp sJolJpJ JspeleJ
I
sselJ aqJ
'apls
,€
eql uo seseq
aql
^q
pJJuJnlJur
sr suopoJ
1a8re1
rraql azruSo
-JJJ
sJoDeJ
IerJepeq
eql
qJrqM qt1a,r
trruaDr;;a
JqJ
EUr
pJIIeJ
,rolJpJ
aspaleJ
I
ssplJ a18urs e
.41uo
sr JJaqt
'sato^rc>lnJ
uI'suopoJ relnrured
roJ sarlrJrlrJads
qlr,u
srolJpJ omt
olur
pa^lola
relel
qlrqm
'suopoJ
uorlpulurJel
11e
pazruSora.r
leql
Jope} eseelal alSuIS
e ,{po se,vl Ilqeqord
aJJql arull
Juo
tv
'seurosoqrJ
ual 'tad
dU
euo ol
lualenrnba
'11ar
rad
qJpJ
Jo
selnJelou
009-
JJe
arJql
lsrolJpJ
uope8uole Jo uopeppl
upqt
sle^al
JJMoI
qJnru
te tuasard
aJe
dU
eqJ
'alrs
d
aq1 ur
YNdl-t[ppdadr(1od
arnbar
pue
rlls
Y
ptuosoqlr
aqt
tp tlp
srolJpJ er{J
'vVO
pup
V)n
sezluSoJar
ZIIU
igVO
pup
vvn
sazruSolar
14g'saruanbas
tuJJaJJrp
ro; rr;nads aJp sJotJpJ asealal
I
sself, omt
'!ln 'E
uI
'(JU)
sJolJBJ aspala'r
1
ssep z(q
pazuSoJJr
are
suopoJ uorleu[uraJ
('apor
rrlaua8
aql
Jo
uonnlona aqt sDeuar sgp
Llqe
-unseJd
'aruanbas
taldrrl
e arrnbal
ppoqs
rr,{q'r.
uospJJ
relnrqred ou aq ol sruJes
JJJqI
os
'uop
-ruSorar
uopoJrlup-uopo) uo
puadap
tou
seop
uor1leal aq1)
'sropey
uralord ,{.q
dlparp
pazruSo
-)er
Jre
pue
suopoJ Jeqlo ruorJ
Jauueu
luJJJJJrp
u(1a;qua up ur uortJuny,{aq1
'VNU1
e
dq
paluas
-ardar
sr suopoJ uorlpunuJal aql
Jo
euoN
'slrunqns
slr olur
auosoqrJ
aql
Jo
uoIlPIJossIp
puP
YNutu
puP
yNul
eql
Jo
JSPJIJJ
sa^Ionul u0q)oaluqtluutl.uJal
-6od
aq1
'VNUI
tsel
Jql ruoJ] uleqJ utalord aql
JO
aSPJIJJ
SaAIO^UI
llesll
U]tpoal
u0tloutwJal erll
'uorlelsupJl
Surpua
uI
pallonul
arB sa8els o,l4l
'(lollPJ
aspolar
I
sspll auo [1uo e,req
qrtqm)
selo&e1na
pue (srope;
aspalol
I
ssel]
]o
sadfl
otnl eneq
qrrqm)
euapeq
ur lepurrs sr. ursruptlrau all
r
'dI9
u0
puaoap
lPql
slopeJ asealal
7
sselr [q
palsrsse
are
sro]lpJ asealar
I
sspl] aql
.
'a6e1u11
y1191
-apqdadr\1od
aq1 azfilorpAq
pup
suopo] uotlputulal
rgrrads o1
puodsar srolleJ asealar
t
ssell aqt
o
'
g-ll pup
n1-13-y11X11\reou
lup
alquasal
slolleJ asealal
I
ssql
eql
Jo
saln]lnlls all
o
'sy11p11freourue
r{q
1ou
's.lollp}
aspelal
uralord
r\q
pezluEorar
alp
suopol uoeeurullOl
r
sjopel
uralojd
Aq
pazruborag
ary
suopol uorlPutujal
srsaqtu^s uralord
8
ulldvHl
,Ll
'uorlerJosseer
Jraql
lueAJJd
01
'asJno)
Jo
'^JessJf,Ju
suIpuIJJ
€-CI
'pJleJedas
arreq
slrunqns Jql JJuo
'lgnqns
s0€
Jqt ruorJ
yNul
patel.dreap
aAourJJ 01 slJe
€-{I
pue
'tlunqns
s0E
Jql uo slJe
cuu irolJPJ
uorlerJossrp
e Jq
o1
pasodo,td
sem
1r
ueqM ,{.raaorsrp
leur8uo
s1r
ot rlrrrr
IInJ IrrqM
Jqt s8uuq rlllqM
'parrnbar
osle sr
€-JI
'uor8ar
Surpurq-poe
ourrue,€ JrIl JoJ
luapnrnba
ue s>lJpl
t1 leql tdarxa
'yNUl
sf,rrurru
lpql
alnlf,nlls
P sPq
cuu
'JSPeleJ
uI
pJAIOAUI
srot)pJ raqto
eql roJ sV
AJg
Jo
srsdlorpLq
sesn
lpql
uorl)eJr
p
ul
t-{g
qtra,r
raqlaSot
sDe
dUU
'(gUU)
rotJeJ SurpLrar rruosoqrJ
sarrnbar
(slrunqns
S0S
pue'S0€
'vN5tu'yNUl)
sluauodruol Surureruar Jql
Jo
uorlerJossrp
eq1
leql
sMoqs
:ri'ii
ri!i;::!:r,
'auosoqrJ
aql
qlrM
prlerJosse
IIIIs
VNUIU
Jqt
pup
VNUI
petelzbe
-ep
p
sJ^eJI
1nq
'apr1dad.{1od
pataldruoJ
eql
Jo
esEJIeJ SaAIOAUT uorlJPJr
UOrleururJl JqI
'zi'1luanry;a
.{.ran
saluanbas
ta8ret
laqt ezruSo
-)aJ
sJolJPJ JSPeleJ JqJ'SUOpOJ
UorleururJl Jql
azruSolar .dlsnoauoua
lpqt
svNUt-1/:eourue
ql1m
saladruor
ZdU
ro
IdU
Lq
uorlruSorar
uopo)
teqt
stsaSSns srql'sDe
tl
qrrqM
uo suopoJ
eql
lp
uoueurruJet
Jo,{ruanrl;a
Jqt saseaJJur
Z{U
Jo
IdU
1o
uorssardxara^O
'uopoJ
uorleururlal
aqr
lsed
srsaqtuLs
uralo,rd anurluo)
ol
.{rrlrqe
pJseJrlur
ue zlq
uaas se
'uorleultural
1o
zi.ruaD
-rJle
eqt aJnpeJ sauaS
gy
eql ur suortetnw
'(s>lJoA\
Jel
-ue)
JSeJJJsuerr
lLprtdad
Jql Moq
Jo
uorssnJsrp
roJ
8t'8
arn8r4 aas)
puoqyNUr-aprtdad
aql
Sur
-2,(1orp.{q
LlaArpJIJJ
snqt leteM aqt
ruor; dno.r8
1z(xo.rpz{q
e sJJJSueJl
uor}pururral
'uorlJeJJ
JJJ
-suerl
aprldad
pnsn
eq1
qlrM
uorpeeJ uorleurur
-
JJl aql saredruor'F
I' r.r
ij
:j
i-1
t i:j'
uorlJPJr JeJsuel]
I{plldad
eql ur
VNgI-1,{reounue
Jo
pr)p
ourrue
Jql JoJ Jlnlrlsqns
ot JInrJIoru Jalem
e uot]rsod
ol
(b)
aunue1n18
rqt asn ol
tl
suolusod slqt
'VNul-l.dreoulurp
up
uo
prJe
ourure
ue
Jo
uorl
-eJol
Iensn
aql ol spuodsJJJo)
Jlrs
V
aql ur uorl
-rsod
sU'Z ureruop;o
do1 aqr
1e
pasodxa
sl
'OCC
'sprJe
oullup
aarql
Jo Jrtoru IerlusssJ
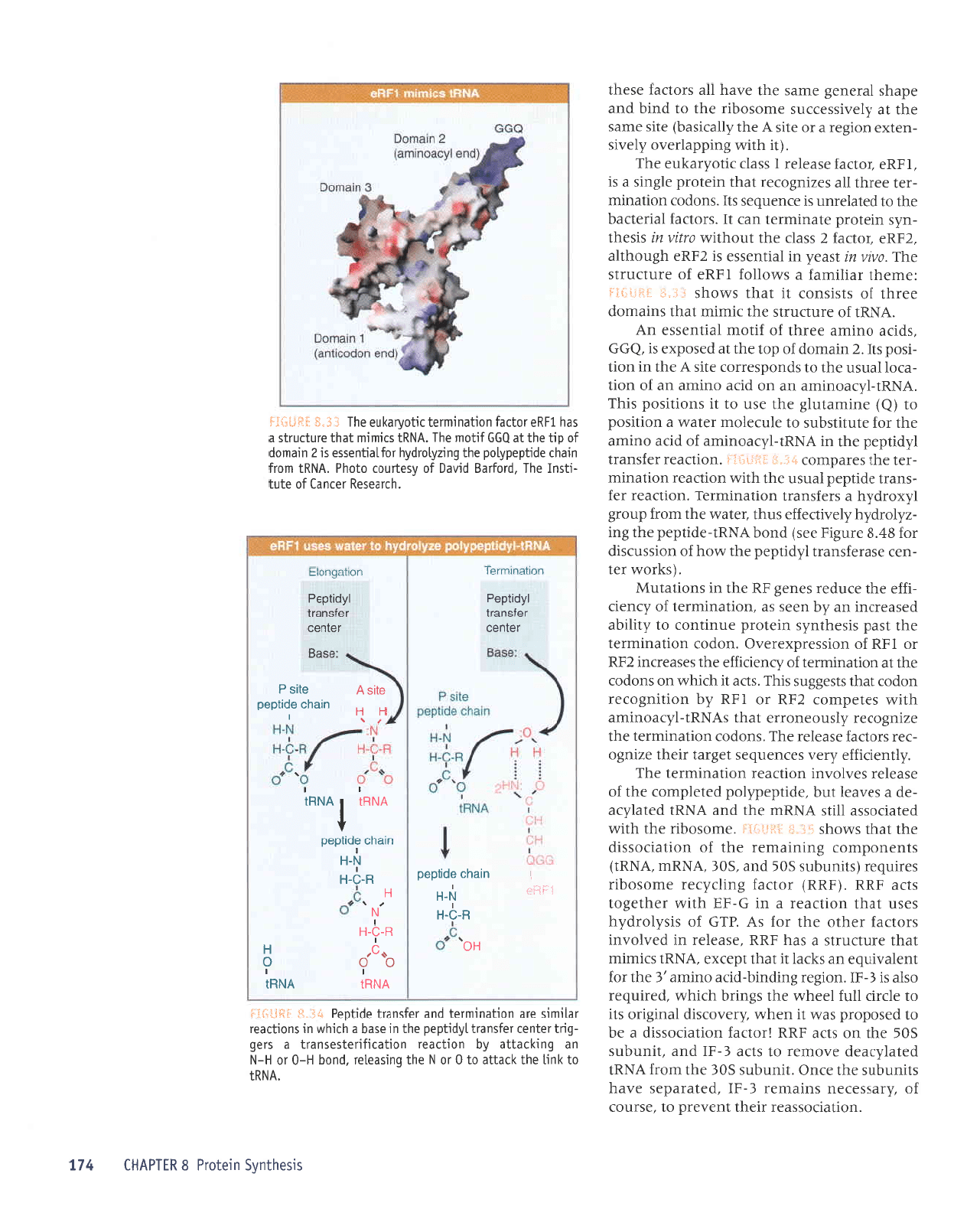
uv
'vNu1
Jo
sJnlJnrls
eqt Jrrulu
lPql
sureuop
aJJql
Jo
slslsuoJ
1l rPql
sMoqs
ri'i:
-liji!:]i.j
:eueql
JPrlIluPJ
p
s/!\ollo]
I{uJ
Jo
JJnt)nJls
etfl
'0^t^
w
tsea(
ur
lerluassJ
sl
Zdua
q8noqrle
'zdu3
'JolrPJ
z
ssPIJ eql
lnoqlrM
0J11^ ut
s$etql
-uLs
uralord
eleurrural
ueJ
lI
'slol)pJ
IerJaDpq
aql 01
pJlelJrun
sr aluanbJs
sl1
'suopoJ
uoueurur
-Jel
JJrql
11e
sazruSotal
leql
urJtoJd
a18urs e sr
'IdUJ
?olJpJ eseJIJJ
I
sselJ llo,{re>lna
aqJ
'(tt
qllr,r
Surddepano Llarrs
-uaJXJ
uor8ar e ro
elrs
y
aqt.dlletrsee) atls
arues
eql
le
,{.lanrssalrns
aruosoqrJ
aqt ol
purq pue
adeqs
leraua8
arues
eql JAeq
ile
sJopeJ
asar{l
.VNUl
ol
)url
or11
llPllP
01
0
lo
N
eql bursealaj
'puoq
H-0
lo
H-N
ue 6ur4re11e
r\q
uorlrear
uorlelrJualsasuerl e slab
-6u1
raluer ralsuerl
lApLldad
aq1 ur aspq
p
qlrr.lM
u! suollleel
rPlrurs eie
uorleurulol
puP
laJsuPll opqoad
bi's:iijtl:lij
'qlleosau
roluPl
Jo
alnl
-qsul
aql
'pjo#eg
ptnp6
Jo
Asalnor oloqd
'VNU1
ulo.U
uteqr apqdadfilod aq1 6urzfi1orpr\q rol
lequassa
sr
Z
urpruop
:ro
0q eql
lP
099
j!10rx
aql
'vNUl
slrulru
leql
arnllnrls P
spq
IlUa
rollpJ uorlpurulal lqo{re1na aq1
i
g
S +d*i};j
url
I
H-C.H
I
N.H
I
ureqc eprlded
VNU}
I
oo
oo'
I
U-C-H
VNHI
I
o
H
i.iu.+
I
N
H
H.C-H
I
N.H
I
ureqc
epuded
A
I
I
VNB}
I
VNU}
tl
\
;r
ureqc aprldoo
a]!s
v
alls
d
:ase8
reluac
ralsuerl
llprldad
uollEUrrnra_L
roluoc
rolsuErl
lIprlde6
uorleOuol3
sLl
slrunqns
leuosoqru
qlog
sape^rad
vNU
leulosoqlu
gI.g
Surpurq
Jeqleqar
Surururalap
u1
tuelrodrur
sr
VNUr
rqt
Io
uoueruroJuot
erp
1pq1
slsa88ns sq1
'punoq
eneq surJtoJd
,raqlo JJIJe
purq
upJ
tnq
'vNur
rrrJ
ol
purq
lou
op srer{lo
'vNur
pJlplosr
o1 .r(18uorts purq
sureloJd
leruosoqrr
aruo5
.YNUI
S€Z
JIIOAJE{OTd
AqT
JO PUJ
,E
Jql
o1 spuodsar:or
aruanbas
stl'vNlI
Sg'g
Jqt
sr
sIqJ
'trunqns
a8rel
eql ur
luesaJd
sr
ypg
leus
rJqloup'sJtuosoqrr
:rlosolA: tuo,{re>1na u1
'erntJnJts
parred-aseq
{U3tq
e
,{eldsrp
salnr
-elou
VNU
SE
IIV'svNUr
roleru rql
Jo
rsoql
ueqt
pJArJsuoJ
IIJ&I
ssJI
sr
VNU
gg
;o
aruanbas
aq1
'(eupuoqJotru
Jo
asoql
ldatxa
seuros
-oqrr
IIp
ul)
17NU SS
eseq
0ZI
e
Jo
alnrrloru
p
surpluoJ
oslp
lrunqns
lpuosoqrJ
a8le1
aq1
'
(erJeDeq ur
palPIAqlJlu
uoruodord
Jql sJrurl aarqt
tnoqe)
pate1.{qtaur
Jre
saprtoelJnu erlt
p
.kZ-
os
',{lanrtradsar
'sdnor81z(qrau
VLpue
gp
ltrct sVNUr
S8Z
pup
s8I
Jql
'sllJ)
uprlPuureru uI
'vNur
sf.zq
0z-
pue (a1nra1ou
rqt
Jo
pua
,€
Jql
preMol
,lltsoru
patelol)
VNUr 59I
ur sdnor8
1[qlaru
0I-
arp
eJJqJ
'sanprsar
patel.dqtJu
Jo
aruasard
aql sr
yNUJ
Jo
arnlJnrts Lreulrd
eql
Jo
ernteal
V
'aler3ossrp
o1 auosoqu aq1 6uLsnet
tUU
sospalol
9-Jl
pup
'VNUI
lspl
eql saspala.l (rope1
6uqrfrer auosoqu)
IUU
aql'ureqr uralotd
aq1 6ur
-sealarfqsrsaqlufsuLelotdsaleurural(rope1
aseelat)
lUoq1
,,.:'ir1
:illrli.,j
'srsaqlu[s
uralo.rd
Suunp alqrxalJ
sr uorleruJoJuoJ aruosoqrJ
tpql
sl
lurod
urpru aql'aJnDnJts
JruosoqrJ ur sa8ueqr
raqlo dq ,(lDarrpur
pesneJ
eJe sJeqlo searaqm
'YNUI
ro
VNUIU
qll^t
YNUr
Jql
Jo
uollf,errlul
lrJrrp
e
DalJaJ
sa8ueqr
rruos
'punoq
sI
vNul
uJqM
Jo
'JterJosse
slrunqns aql ueqM
'punoq
sr
VNUrU
uequ
JnJJo
VNUJ
er{l
Jo
zhrnurpJJ Jql ur
sa8ueq3
'srsaqluLs
uralo:d ur
paSe8ua
asoql
pue
sJruosoqrJ earJ uJJMlaq
SJJUJJJJJIp aJP aJJql
osle
lsJruosoqlJ
S0l,
qtlm parcduroJ
Jrp slrunqns
S0€
uJqM
punoJ
are slua8e
lpJrueqJ
qll,rlr
ppeJ
ol
VNUr
59I
Jo
^tqlqp
aql
ut srruereJJr(
'lrunqns
eqt ur
uorlpJol eleJJsrp
e a^eq
pup,{lluapuadapq plol
ygg.r
roleru aql
Jo
sureuop
eql
lunqns
qJea
ut
leql
sMoqs
auosoqlJ Jql
Jo
aJntJnrls
plsLn
JqJ'sureuop
Ipuontppe
Surluasarda; saruanbas
;o
uorlrsrnbJe eqt o1
.{1a8re1 Jnp sI syNUr
:rlodre>1na
ur
qfual
uI as€al)ul
aq1
'(suteruop
aJoru JApq
pue
raEuol are
qttq,ror)
sVNUJ
rrloso1z(r
rr1o,{.re1na roJ
pup
(sureurop
JaMeJ
eleq
pue
JJuoqs are
qrtq,vr)
sVNUJ
IeIJpuorIJ
-olrrr
JoJ umpJp
uJJq e^eq slapou
alqeredruo3
'saseq
parredun
;o
sa8pq ureluoJ
pue
pa;rad
lou
erp
suor8ar xaldnp aqt urUO
'(dq
g>)
uoqs
aq 01
puJt
suor8ar
IeJrlJq-alqnop
lenpl^tpul
eqJ'VNUr
S€e
(qpeuroJ
erp sulpruop
leraua8
xls
'(Et'8
arn8r4
aas)
parred
aseq sI aruanbas
ar{l
}o JIeq
rrpun
lsnl
qttq,rn
ul
'VNUr
S9I
zi.q
paruro;
eJp sureruop
praua8
Jnod
'sulpruop
aleJ)srp
IeJJAJS
qll^{
JJnllnJlS
^JepUOJJS
P
ur uMeJp Jq ueJ
sVNUJ ;o[eu eql
Jo
q)ea
'YNU]
S€Z
pue
S9I
qloq
JoI
prlJnrlsuoJ
Jq
01 slepolu
pellPlep
pJlqeua
seq
qreo.rdde
stql
'yNUr
qJpe
ur uorlrsod
Jlrlelal
Jrues Jql
le
IUJoJ ue)
ll
'parrnbar
sr
rred Jseq e
]l
snql
'Sutrted
aseq
dq
lJeJJlur
o1
,{1r1qe aq1 urplJJ arntf,nJts
Lrepuo
-Jas
eqt ur
tuelrodurl
aJe
1eq1
suot8ar esoql
'srusrue8ro
pelplJJ
uI syNuJ
Sutpuodsarror
;o
saruanbas aql
aredruoJ ol
sr sypg a3;e1
yo
arnlJnJls,{.repuoras
Sutzz{.1eue o1
qreordde
8ut
-terlauad
tsoru
aqJ
'VNUJ
yo
dn aperu sI etuos
-oqrr
IPrrJlJEq
aqr
Jo
ssPlu eqr
Jo
sprlql o^\L
'svNul
s€z
puP
s9l
eql uaaMlaq apeu are
slrunqns
lPurosoqu
uaaMlaq
slleluol aql
J0
lsol^|
.VNUI
qlrm
lleluol
ur are sulalord
leuosoq.u
1e
411en111
'Alluapua0apur
ploJ
lPql
sureurop
]lutlstp
lPle^as
sPtl
vNUl
qrel
slrunqns
lPruosoq!u
qroB
sapPruad
vNU
lPl.uosoqtu
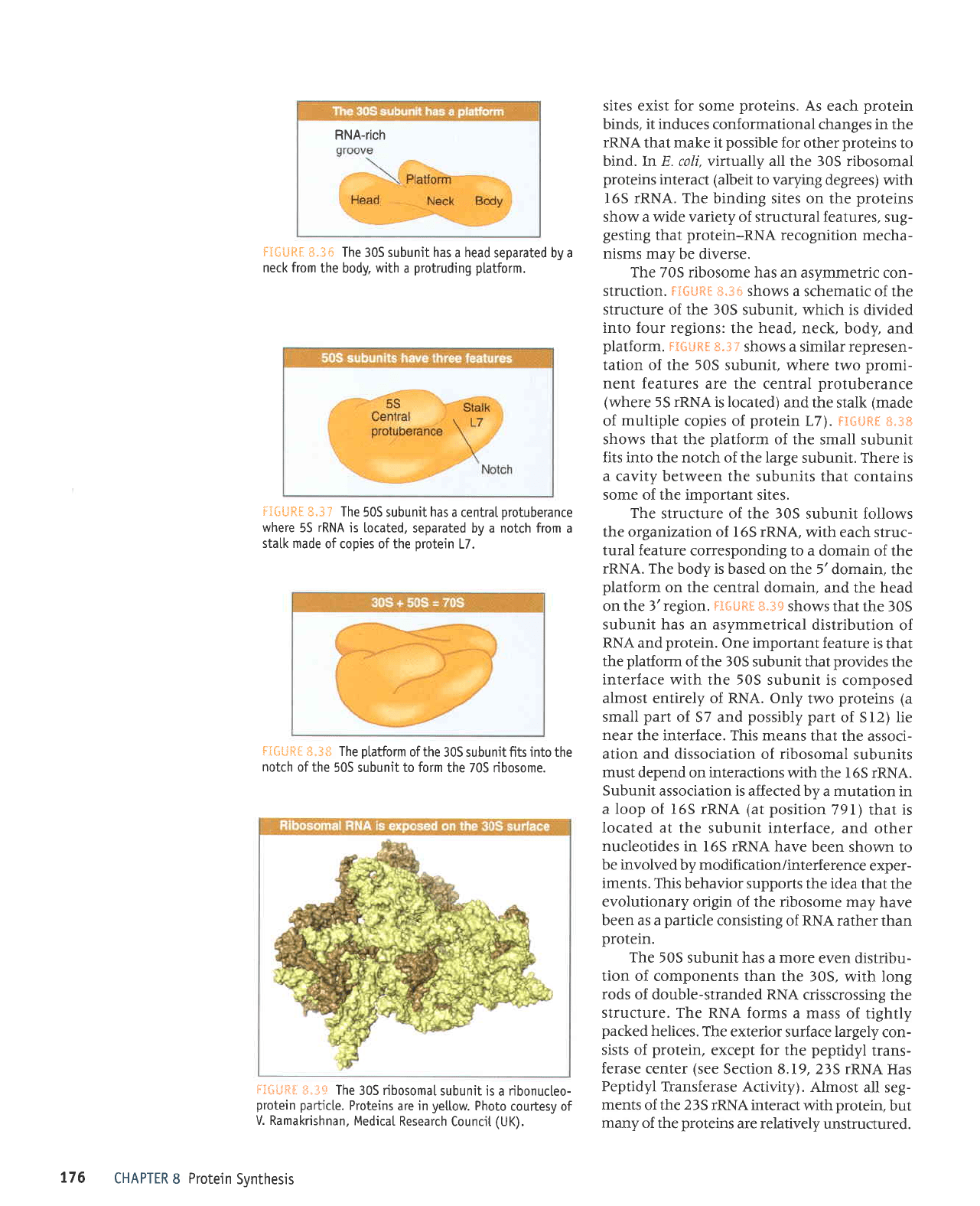
RNA-rich
FI6URI
S.36
The
30S subunit has
a
head
separated by a
neck from
the body.
with
a
protruding ptatform.
sites
exist
for
some
proteins.
As
each
protein
binds,
it induces
conformational changes in the
rRNA
that make it
possible
for other
proteins
to
bind. In E. coli, virtually all the 30S ribosomal
proteins
interact
(albeit
to varying degrees)
with
I65 rRNA. The binding sites
on the
proteins
show a wide variety
of structural features, sug-
gesting
that
protein-RNA
recognition
mecha-
nisms may
be diverse.
The 70S ribosome has
an asymmetric con-
struction.
FISIJftil
*,3& shows a
schematic of the
structure of the l0S
subunit, which is divided
into four regions:
the head, neck,
body, and
platform.
FIGUR{
*.3? shows a similar represen-
tation of the 50S
subunit, where two
promi-
nent
features
are the central
protuberance
(where
55 rRNA is located)
and the
stalk
(made
of multiple copies of
protein
L7). FI*Ufi{
s.3*
shows that the
platform
of the
small subunit
fits into the notch of the large
subunit. There is
a cavity
between the subunits that contains
some of the important
sites.
The
structure of the 30S
subunit follows
the organization of l65 rRNA,
with each struc-
tural feature
corresponding to a domain
of the
rRNA. The
body is based on the 5'domain,
the
platform
on the central
domain, and the head
on the
3'region.
F:fitJRC
S.3$ shows
that the 30S
subunit has an asymmetrical
distribution
of
RNA and
protein.
One important
feature is
that
the
platform
of
the l0S
subunit that
provides
the
interface
with the
50S
subunit is
composed
almost entirely of RNA. Only
two
proteins (a
small
part
of 57 and
possibly part
of SI2) Iie
near
the interface. This means
that the
associ-
ation and dissociation
of ribosomal
subunits
must
depend on interactions
with the I65 rRNA.
Subunit
association
is
affected by
a
mutation
in
a
loop
of l65 rRNA
(at
position
791) that is
located at
the subunit interface,
and other
nucleotides
in l65 rRNA have
been shown
to
be involved
by
modification/interference
exper-
iments.
This behavior
supports the idea
that the
evolutionary
origin of the ribosome
may have
been as a
particle
consisting
of RNA rather
than
protein.
The 50S subunit has
a
more
even
distribu-
tion of components than
the 30S,
with Iong
rods
of double-stranded
RNA crisscrossing
the
structure. The RNA
forms a mass
of tightly
packed
helices.
The exterior
surface largely
con-
sists of
protein,
except for the
peptidyl
trans-
ferase
center
(see
Section 8.19, 23S
rRNA Has
Peptidyl
Tlansferase Activity).
Almost
all seg-
ments
of the 23S rRNAinteractwithprotein,
but
many
of the
proteins
are relatively
unstructured.
FIfiUR[
$.3? The
50S subunit
has
a centraI
protuberance
where
55
rRNA
is located,
separated by a notch from
a
statk made
of copies of the
protein
17.
FiGriR[
8.3$ The
ptatform
ofthe
30S subunitfits into the
notch
of the
505 subunit to form
the 70S ribosome.
Fitu#Rg
*.3# The 30S ribosomal
subunit is
a
ribonucteo-
protein
pariicle.
Proteins
are in
yetlow.
Photo
coudesy of
V. Ramakrishnan.
MedicaI
Research
Councit
(UK).
CHAPTER
8
Protein
Synthesis
176
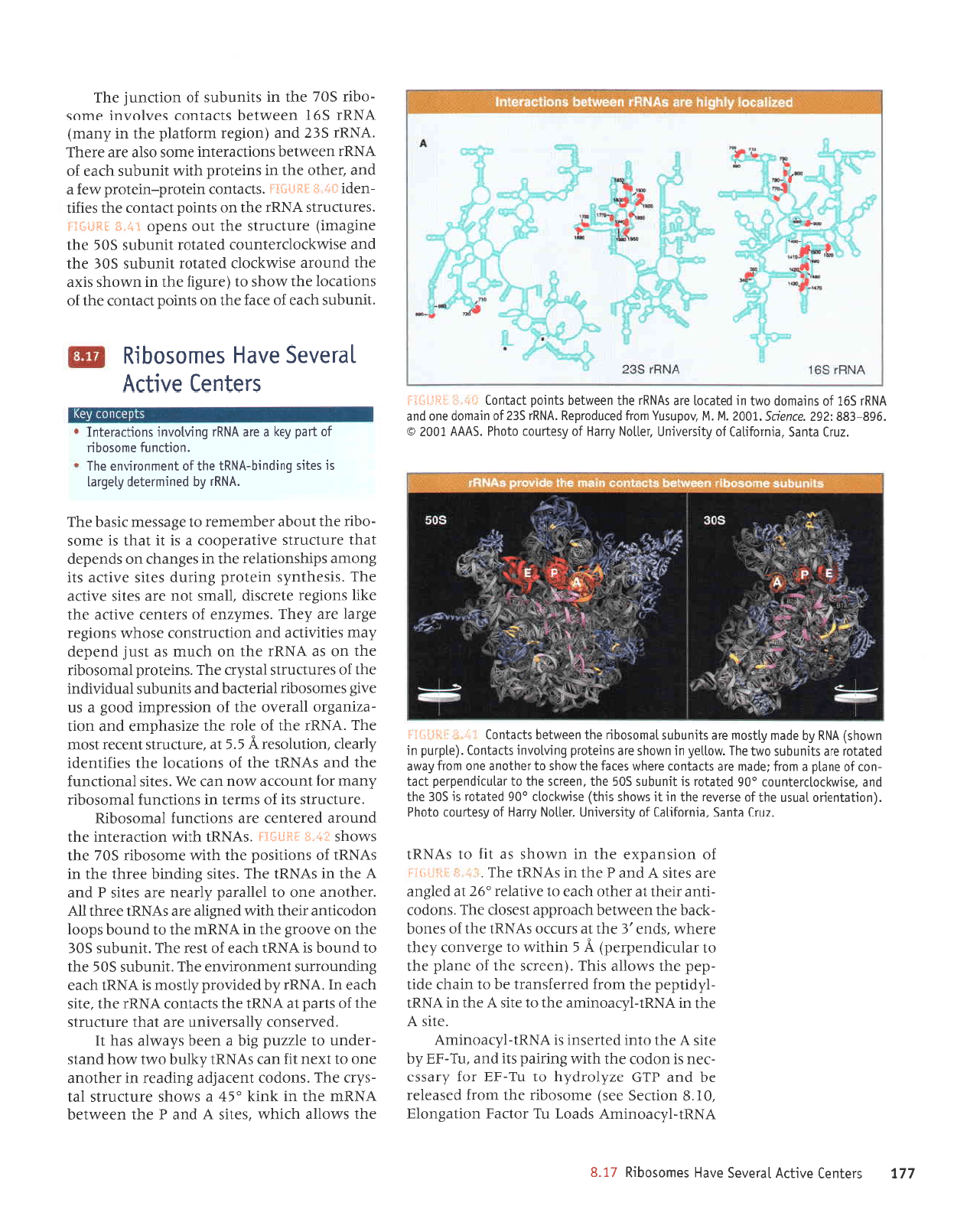
LLI
sralual a^L]lv
lpra^as
o^pH seuosoqru
/I'g
YNUI-lrbeouruY
spPol
I\l
JoDed uotle8uolg
'0I'8
uortJJ5
aas) aruosoqu
Jql uoJJ
pJSeJIJT
rq
pup
419
az.dlorpAq
or nJ-{iI ro; Lressa
-Jeu
sr uopor Jql
qlrM
Suured sll
pue
'nI-{g
^q
atIS
V
Jql otur
peuasur
sr
y1qgl-1trrpoq1v
Jqr ur
vNut-lLreoutue
eqt ot Jus
O
,q,
";?ilJ
-llpttdad
aql
uoJJ
pJrreJsuert
rq ot ureqr epu
-dad
aqt s.tr.olle srql
'(uaans
Jq1
Jo
aueld
aql
ot relnJrpurdrad)
y
E
urqlrM o1 a8ranuor.{aql
arJqM
'spuJ,€
Jql
le
sJnrJo svNul eql
Jo
sJuoq
->lJpq
rqt
uJeMlJq
qJeordde
tsasop
JqJ
'suopoJ
-Itup
Jreql
lp
rJqto
qJee
01 anrleleJ
"9e
rc
pelSue
eJe SJUS
V
pue
d
eql uI sVNUf a{J
'l
r.
lr
.li.'rii:-11:i
;o
uorsuedxJ Jqt
ur uMoqs se
lrJ
ot sVNUl
'znll
eluPS
'eruloJrlPl
1o
rtlrstanrul
'ta11o1t1
rfueg
;o
Asalnor o1oq6
'(uorleluauo
lensn
aql
Jo
as.la^el eql
ur
1r
sMoqs srql) asLr'r1ro1r
o06
palplol
s!
S0E
aql
pue
'asrmllollrolunol
o06
palplol
sr
ltunqns
S0g
oql
'uaells
or.1l o1 telnrrpuadtad
1re1
-uor
J0
aueld
p
ruo.lJ
loppu
ale slleluol olotlM saleJ
oql Moqs ol.laqloue auo uor;
ferne
palplor
olp slrunqns oMl
aql'ano11a[ ur umoqs ele
suralold 6uurlonuL sl]pluol'(aldtnd
ur
ur'roqs)
ylx
fq
apeu [11sou
are slrunqns
lprxosoqp
aql uoaMloq sl]eluol
1
1' r
l,r{:a}Lj
'zn.ll
plups
'erulo1qp3
1o
filstenrug
'ra11oy
fueg
1o
fsalnor
oloqd
'SVVV
t00Z
o
'968-E88
iZ6Z
'auaDs'IOOZ
'14 '14 'nodnsnl
uo4
pernpotdau
'VNUI
SEZ
Jo
urpurop
euo
pup
VNU.I
59l
Jo
surpulop oMl
ur
palelol
ale
sVNUr aql uaamloq sluLod
1re1uo1 .,"
i;l;1:t+lj
eqt sA1.ollP
qJIqM
'sJlrs
Y
pue
d
aql uaaMlaq
vNuru
eql
ur
{ul>l
.gt
e sMoqs
arnlJnrls
Iel
-sLn
aq1
'suopo)
tua)elpe
Sutpea; uI Jeqloue
euo 01
txeu
tlJ
ueJ sVNUt,{11nq oanl
llroq
puPts
-rJpun
o1 alzznd 8rq e
uaaq sleule seq
U
'pJAJaSUOJ
LlleSramUn AJe
leql
alnlf,nJls
aqt
yo
sued
1p
VNut
aqt stf,eluoJ
vNUr
eql
'a1ls
qreJ
uI
'VNUJ
z(q
papnord
[llsour sr
VNUI
qJpe
Surpunorrns
tuauruoJrlua
JqJ'trunqns
S0E
Jql
ol
punoq
sl
vNur
qrea
Jo
lsar
aqJ
'llunqns
s0€
Jqt uo anoor8
Jql uI
VNUIU
aq1
ot
punoq
sdool
uopoJrtup Jraqr
qryu pau8rtp
Jrp sVNUI JJJqI
IIV
'raqloup
Juo 01
1a11ered
ui.peau JJp sJlIS
d
pue
v
aql ur svNul
eqJ
'selrs
Surpurq
aerql eqt uI
sVNUt
yo
suorlsod eqt
qtl,t.t
JruosoqlJ
S0Z
eql
sMol{s
;j+'g
-+}lil:l]..
'sYNUl
qll1\{
uolperelul
rql
punoJe pJraluel
aJe suorlJunJ
Ieluosoqlu
'eJnlJUlS
SU
JO
SruJJl UI SUOIIJUnJ
IPTUOSOqIJ
,{.ueru ro;
lunoJf,e
Mou upJ a714'sells
Ieuollf,unJ
rql
puP
svNur
aql
Jo
suollef,ol
eql srlJlluapl
z(1reap
'uorlnloseJ
y
E'S
tp
'JJnpruts
IUJJJJ lsoru
aqJ
'VNUJ
aql
Jo
JIoJ eqt aaseqdruJ
pue
uoll
-ezruv?to
ilerJno
eql
Jo
uorssa:drur
poo8
e sn
arrrS
saurosoqrJ
IPrJapeq
pue
sllunqns
pnpIAIpuI
aql
Jo
sernDnrls
plsrlD
aq1
'sutalord
leluosoqlJ
eql uo se
vNur
rql uo
qJnru
se
1sn[
puadap
.,i.eru sartrnrlre
pup
uollf,nJlsuoJ
asoqm
suor8ar
a8rel are
daql
'saru,{zue
lo
sreluJf, a^IDp eql
a111 suor8ar
eleJf,srp
'lleurs
lou
aJp sJtls
eAIlJe
JqJ
'srsJqlu.ds
uralord Surrnp
sells eAIlJp sU
Euorue sdrqsuoqelal
eql
q
sa8ueqr uo spuadap
leql
eJnlJnrls a,rtleradooJ
P
sI
lI lPrll
sI Jruos
-oqlr
aql
tnoqp
JJqIUJTUJJ
01 a8essaur
rtseq aq1
'VNUr
fq
paurutalap
r{1a6re1
sL salrs 6urpurq-VNUI
aql
Jo
luotlluolt,rua
aqj
u0qrunj
euosoqu
;o
1ed
r\el
p
arp
VNUI
burnlonut
suoqlelalul
srolual
aAqlv
lue^as
a^eH
sauosoqtu
't1unqns
q)pa
Jo
a)eJ
Jq1 uo stutod
peluoJ
eqt
Jo
suorlef,ol eql
Moqs o1
(arn34
eql ul uMoqs
sIXe
eql
punoJP
asrM>lJolJ
pelelor
llunqns
s0€
aql
pue
Jsr!\>lJolJlalunoJ
pJlelor
llunqns
s0E
eql
aur8erur)
arnlJnJls Jql
lno
suado
1t'E
SEfttl'j
'seJnDnrls
VNUJ
eqr uo
slurod
lJpluoJ
Jql
SJIJI1
-uJpI
{.iir"iq .+s*1i;-{
'slJpluo)
utatord-utalord
ma; e
pue
teqto
Jql
q
sulJlord
qtt,u
lpnqns
I{JeJ
Jo
vNuJ
uJeMlaq
SuOrlJ€rJluI
aruos oslP ale aJaqJ
'vNUr
S€Z
pue
(uorSar
ruropeld
aql ur dueur)
vNur
s9I
uaaMlaq
slJeluof,
seAIoAuI auros
-oqlJ
S0L
Jqr ut
sltunqns
;o
uortrunI aq1
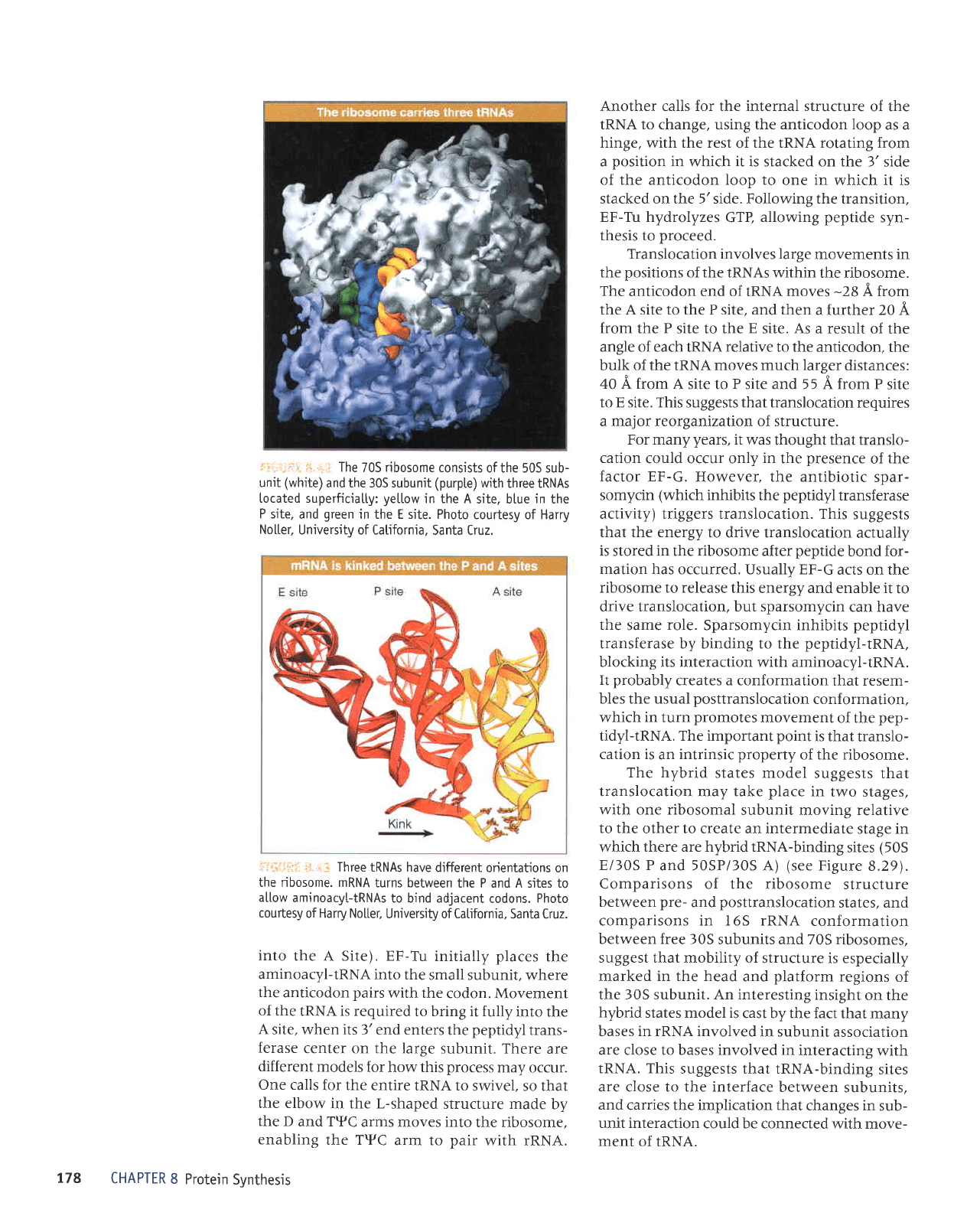
:.:::;j-.:::
.::.r,,i
The
70S
ribosome
consists
of the 505 sub-
unit
(white)
and the 305 subunit
(purpte)
with
three tRNAs
located superficiatly:
yettow
in the A
site, btue
in
the
P site,
and
green
in the E site. Photo
courtesy of Harry
No[ter,
University of Catifornia,
Santa Cruz.
i:r.iii.ir
r1..l'
:
f[1gg tRNAs have
different
orientations on
the ribosome. mRNA
turns between
the P and A sites to
a[[ow
aminoacyltRNAs
to bind adjacent
codons.
Photo
courtesy
of Harry No[[er,
University of CaUfornia.
Santa Cruz.
into
the A Site). EF-Tu
inirially
places
the
aminoacyl-IRNA
into the
small subunit, where
the
anticodon
pairs
with the
codon. Movement
of the
IRNA is required
to bring it fully
inro rhe
A
site, when its
3'end enters
the
peptidyl
trans-
ferase
center on the large
subunit. There
are
different
models
for how this
process
may
occur.
One calls
for the
entire 1RNA to
swivel, so that
the elbow
in the L-shaped
structure made
by
the D
and TYC
arms moves into
the ribosome,
enabling
the TYC
arm to
pair
with IRNA.
Protein
Svnthesis
Another calls for the internal structure
of the
IRNA to change, using the anticodon loop
as a
hinge, with the rest
of
the IRNA rotating from
a
position
in which it is stacked on the J'side
of the anticodon loop to
one
in
which it is
stacked on the 5'side. Following the
transition,
EF-Tu hydrolyzes
GTP,
allowing
peptide
syn-
thesis to
proceed.
Translocation involves large
movements in
the
positions
of the tRNAs within
the ribosome.
The
anticodon end of IRNA moves
-28
A from
the A site to the P site, and then
a further 20 A
from
the
P
site to the
E
site. As a result
of the
angle of each IRNA relative to the anticodon,
the
bulk of the IRNA moves much larger
distances:
40
A
from
A site to P site and 55 A from P
site
to E site. This
suggests
that
translocation requires
a major reorganization of structure.
For many
years,
it was
thought that translo-
cation
could occur only
in
the
presence
of
the
factor
EF-G. However, the antibiotic
spar-
somycin
(which
inhibits
the
peptidyl
transferase
activity) triggers translocation. This
suggests
that the energy
to drive translocation
actually
is
stored
in
the
ribosome
after
peptide
bond for-
mation
has occurred. Usually EF-G
acts on the
ribosome to release
this energy and enable it
to
drive translocation, but sparsomycin
can have
the
same
role.
Sparsomycin inhibits
peptidyl
transferase by binding to the
peptidyl-IRNA,
blocking its interaction with aminoacyl-IRNA.
It
probably
creates a conformation
that resem-
bles the usual
posttranslocation
conformation,
which
in
turn
promotes
movement
of the
pep-
tidyl-tRNA. The important
point
is that
translo-
cation is
an
intrinsic
property
of the ribosome.
The hybrid
states model
suggests that
translocation may take
place
in
two stages,
with one ribosomal
subunit moving relative
to the
other to create an intermediate
stage in
which there are hybrid tRNA-binding
sites
(50S
E/30S P
and 50SP/30S A)
(see
Figure
8.29).
Comparisons of the ribosome
structure
between
pre-
and
posttranslocation
states, and
comparisons in 165 rRNA
conformation
between free l0S
subunits and 70S ribosomes,
suggest that mobility of
structure is especially
marked in the head
and
platform
regions
of
the 30S subunit. An interesting
insight
on the
hybrid
states model is
cast by the fact
that many
bases in rRNA involved
in subunit
association
are close to bases involved in
interacting
with
IRNA. This
suggests that IRNA-binding
sites
are close to the interface
between
subunits,
and
carries the implication that
changes in
sub-
unit interaction
could be connected
with move-
ment of IRNA.
778
CHAPTER
8

Much
of the structure
of the ribosome
is
occupied by its
active centers.
The schematic
view of the ribosomal
sites in
'
,.
i
shows
they
comprise about two
thirds of
the
riboso-
mal structure. A IRNA
enters
the A site, is trans-
ferred
by translocation into
the P
site. and then
Ieaves the
(bacterial)
ribosome
by the E site.
The A and P
sites extend across
both ribosome
subunits; IRNA is
paired
with mRNA in
the 30S
subunit,
but
peptide
transfer
takes
place
in
the
50S subunit. The A
and P sites
are adjacent,
enabling translocation
to move
the IRNA from
one site into the other. The E
site is located near
the P
site
(representing
a
position
en route to
the surface of the 50S
subunit). The
peptidyl
transferase center is located
on the 50S
subunit,
close to the aminoacyl
ends of the
tRNAs
in
the
A and P sites
(see
Section
8.18, l65 rRNA Plays
an Active Role in Protein
Synthesis).
All
of
the
GTP-binding
proteins
that func-
tion in
protein
synthesis
(EF-Tu,
EF-G, IF-2,
and RFl,
-2,
and
-3)
bind to
the same factor-
binding site
(sometimes
called the
GTPase cen-
ter), which
probably
triggers their hydrolysis
of
GTP.
This
site
is
located at the
base of the stalk
of the large subunit,
which
consists of the
pro-
teins L7 and Ll2.
(L7
is
a modification
oILl2
and
has
an acetyl
group
on the
N-terminus.) In
addition to this region,
the complex
of
protein
Ll l with a 58-base
stretch of 23S rRNA
pro-
vides
the binding site for some
antibiotics that
affect GTPase activity.
Neither of these riboso-
mal
structures actually
possesses
GTPase activ-
ity, but they are both necessary
for it. The role
of the
ribosome
is to trigger
GTP hydrolysis by
factors bound in the factor-binding
site.
Initial binding
of 30S subunits to nRNA
requires
protein
S l, which has a
strong affinity
for single-stranded nucleic
acid. It is responsi-
ble
for maintaining
the single-stranded
state in
nRNA that is bound to the
30S subunit. This
action is necessary to
prevent
the mRNA from
taking up a base-paired conformation
that
would be unsuitable for translation.
S
I
has an
extremely elongated structure and associates
with S18 and 52l. The three
proteins
consti-
tute a domain that is involved
in the initial bind-
ing
of
nRNA
and
in
binding initiator IRNA. This
locates
the
mRNA-binding
site in the
vicinity
of the cleft of the small subunit
(see
Figure 8.3).
The
3'end of
rRNA,
which
pairs
with the nRNA
initiation site, is located in this region.
The initiation factors bind in the
same
region
of
the ribosome. IF-l
can be crosslinked to the
3' end of
the rRNA,
as well as to several ribo-
somal
proteins,
including those
probably
involved in
binding
nRNA.
The role of IF-3
I
'
,
,.
The ribosome
has severa[ active centers.
It
may
be associated
with a
membrane. mRNA takes
a turn
as
it
passes
through
the A and
P sjtes, which are angted
with regard
to each other.
The
E
site
lies beyond the
P sjte.
The
peptidyt
transferase
site
(not
shown) stretches
across
the tops of the A and
P
sites.
Part of the site
bound by
EF-Iu/G ljes at the base
of the
A
and
P sites.
could be to stabilize
mRNA-30S subunit
bind-
ing;
then
it
would
be displaced
when
the 50S
subunit
joins.
The incorporation of
5S RNA
into 50S sub-
units that are assemble
d
in vitro depends on
the
ability of three
proteins-L5, L8, and L25-to
form a
stoichiometric
complex
with
it. The com-
plex
can bind to
2lS
rRNA, although
none of
the isolated components
can do so.
It lies in the
vicinity
of the
P and
A sites.
A nascent
protein
debouches
through
the
ribosome, away
from the active
sites,
into the
region in which ribosomes
may
be attached to
membranes
(see
Chapter
10, Protein
Localiza-
tion). A
polypeptide chain emerges
from the
ribosome
through
an exit
channel,
which leads
from the
peptidyl
transferase
site to the surface
of rhe 50S subunit.
The tunnel
is composed
mostly
of
rRNA. It is
quite
narrow-only
I to
2 nm wide-and
is
-10
nm
long. The
nascent
polypeptide
emerges
from the
ribosome
-I5
A
away from the
peptidyl transferase
site. The
tunnel can hold
-50
amino
acids, and
proba-
bly constrains
the
polypeptide chain so that
it
cannot fold until
it leaves
the exit
domain.
165 rRNA
Plays an
Active
Ro[e
in
Protein Synthesis
.
165 rRNA
ptays
an active
rote in the functions
of
the 30S subunit.
It
interacts directty
with mRNA,
with the 50S subunit,
and
with the anticodons
of
tRNAs in the
P
and
A sites.
The ribosome was
originally
viewed
as a col-
lection of
proteins with various
catalytic
8.18
165
rRNA
Ptays an Active
Rote
in Protein Synthesis
179
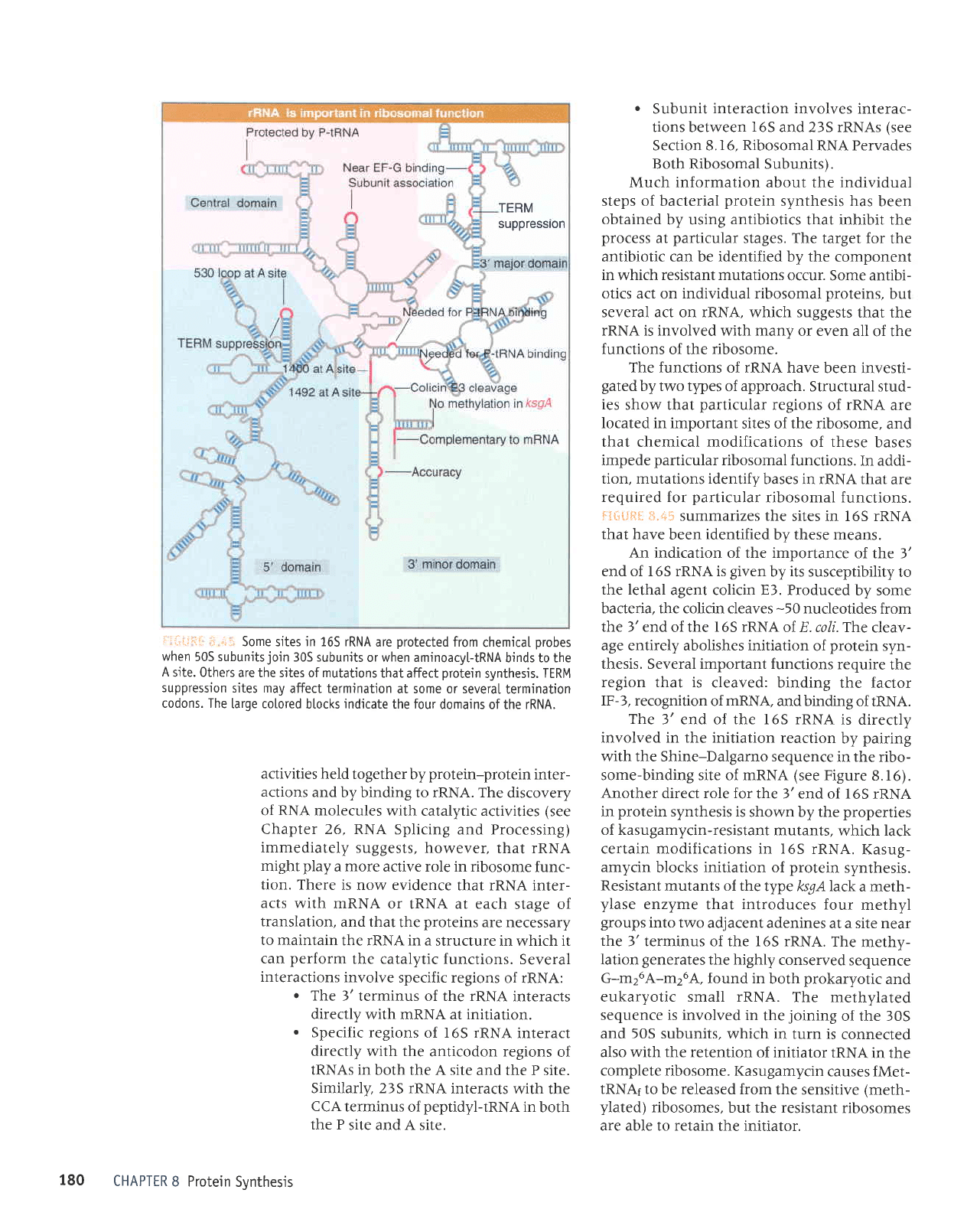
r:i:-:.i:
.:
,i::
Some sites in 165 rRNA
are
orotected
from
chemical orobes
when
50S subunits
join
30S subunits
or when aminoacyltRNA
binds to the
A
site. 0thers
are the sites of mutations
that affect
protein
synthesis.
TERM
suppression
sites may
affect termination
at some or several
termination
codons. The
[arge colored
btocks
indicate
the four
domains ofthe rRNA.
activities
held together
by
protein-protein
inrer-
actions
and
by binding to rRNA. The
discovery
of
RNA
molecules
with catalytic activities
(see
Chapter 26, RNA
Splicing
and Processing)
immediately
suggests, however,
that rRNA
might
play
a
more
active role in ribosome
func-
tion. There
is now evidence
that rRNA inter-
acts with
nRNA
or tRNA at each
stage of
translation,
and that
the
proteins
are necessary
to maintain
the rRNA in a
structure in which it
can
perform
the catalytic functions.
Several
interactions
involve
specific regions
of rRNA:
.
The
3' terminus of the rRNA
interacts
directly
with mRNA at initiation.
.
Specific regions
of l6S rRNA
interact
directly with the
anticodon regions
of
tRNAs in
both the A site
and the P site.
Similarly, 23S IRNA
interacts
with the
CCA
terminus
of
peptidyl-tRNA
in
both
the P
site and A site.
CHAPTER
8 Protein
Synthesis
.
Subunit
interaction involves
interac-
tions between
I65
and 23S rRNAs
(see
Section 8.16,
Ribosomal
RNA Pervades
Both Ribosomal Subunits).
Much information about
the
individual
steps of bacterial
protein
synthesis has
been
obtained
by using
antibiotics
that inhibit the
process
at
particular
stages. The target for
the
antibiotic can be identified by the
component
in which resistant mutations
occur. Some antibi-
otics act
on
individual ribosomal
proteins,
but
several act on rRNA, which suggests
that the
rRNA is involved
with
many
or even all
of the
functions
of the
ribosome.
The functions of rRNA have
been investi-
gated
by two types
of
approach.
Structural stud-
ies
show that
particular
regions
of
rRNA
are
located in important
sites of the ribosome,
and
that chemical modifications
of
these
bases
impede
particular
ribosomal functions.
In addi-
tion, mutations identify
bases in rRNA that
are
required for
particular
ribosomal functions.
Fg*tJftil $.4*
summarizes the
sites in l6S rRNA
that have
been
identified
by these means.
An indication of the importance
of the 3'
end of I65 rRNA is
given
by
its
susceptibility
to
the lethal
agent colicin E3. Produced
by some
bacteria, the colicin cleaves
-50
nucleotides
from
the 3'end
of the
I65 rRNA
of E. coli.The
cleav-
age entirely abolishes initiation
of
protein
syn-
thesis. Several important functions
require
the
region that is
cleaved: binding
the factor
IF-3, recognition
of
mRNA,
andbinding
of IRNA.
The
3' end of the 165 rRNA
is directly
involved in the initiation
reaction
by
pairing
with the Shine-Dalgarno sequence
in the ribo-
some-binding site of mRNA
(see
Figure
8.16).
Another direct role for
the 3'end of 165
rRNA
in
protein
synthesis is
shown by the
properties
of kasugamycin-resistant
mutants,
which lack
certain modifications in l65
rRNA.
I(asug-
amycin
blocks initiation of
protein
synthesis.
Resistant
mutants of the
type ksgAlacka
meth-
ylase
enzyme
that
introduces
four
methyl
groups
into two
adjacent adenines
at a site near
the
3'terminus of the 165 rRNA.
The
methy-
lation
generates
the highly
conserved
sequence
G-m26A-m26A,
found in
both
prokaryotic
and
eukaryotic small rRNA.
The methylated
sequence is involved in
the
joining
of the 30S
and
50S
subunits,
which in turn is
connected
also with the retention
of initiator
IRNA
in the
complete ribosome. I(asugamycin
causes fMet-
tRNAl to
be
released
from
the sensitive
(meth-
ylated)
ribosomes,
but the resistant
ribosomes
are able to retain
the initiator.
TERM
suppressron
180
